Abstract
When natural populations exchange migrants at a rate proportional to their geographic distance, population genetics theory leads to the expectation of a pattern of isolation-by-distance (IBD), whereby geographic and genetic distance are correlated. However, the presence or absence of such patterns in modern populations may not fully reflect the historical relationships among those populations. Thus, historical samples, collected prior to modern human impacts, can often provide a critical baseline for comparison with modern populations. Steelhead, the anadromous form of rainbow trout, Oncorhynchus mykiss, are native to western North America and are endangered or threatened throughout most of California, near the southern extent of their native range. Population samples of steelhead collected in 1897 and 1909 in Central California rivers provided the opportunity to evaluate the historical genetic composition and population structure of these threatened fish. Here we show that these steelhead populations had a historically strong correlation between genetic and geographic distance that has been virtually erased in modern populations, suggesting that current relationships among modern steelhead populations are no longer reflective of natural migratory pathways. This demonstrates the critical role of migration in maintaining population relationships of threatened species and highlights the importance of natural history museums in providing historical baseline information.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In structured populations, migration increases the effective size of local demes and spreads novel mutations/adaptations. It also determines, among other things, the rate of recolonization of newly available habitat and the efficacy of protected areas. Understanding migration is therefore a critical component of species and ecosystem conservation. When natural populations exchange migrants at a rate proportional to their geographic distance, population genetics theory leads to the expectation of a pattern of isolation-by-distance (IBD; Wright 1943), whereby geographic and genetic distance are correlated. However, as has been noted in fisheries and wildlife management (Jackson et al. 2001; Newsome et al. 2007), human-altered systems often do not provide an accurate representation of historical conditions (shifting baseline syndrome; Pauly 1995; Pinnegar and Engelhard 2008). From a population genetic perspective, patterns of genetic variation observed in modern populations may not accurately reflect the historical relationships among their ancestral populations. Recent anthropogenic impacts, through habitat modification or species management practices, have the potential to significantly alter the genetic structure of species. In such situations, inference based solely on data from contemporary populations may lead to inaccurate conclusions about historical population relationships and misguided management decisions and conservation goals.
Natural history museum collections represent a valuable source of direct data from natural populations prior to widespread human impacts, and recent studies have demonstrated the utility of museum specimens as a source of DNA for genetic analysis and comparison with modern populations (Wandeler et al. 2007; Leonard 2008; Nielsen and Hansen 2008). In particular, the ability to obtain DNA from substantial numbers of museum specimens has opened the door to more extensive studies that compare historical population-level genetic patterns with those from extant populations in the same habitats (e.g. Heath et al. 2002; Leonard et al. 2005; Lozier and Cameron 2009). For example, Hansen et al. (2009) used scale samples archived in the mid-1900s to show the effect of hatchery stocking on several brown trout (Salmo trutta) populations in Denmark. Such data may be used to help understand and preserve (or restore) the historical evolutionary processes among species or groups of populations (Crandall et al. 2000).
The species Oncorhynchus mykiss encompasses anadromous and resident fish with diverse life-history strategies, often within a single population, and includes a number of named subspecies and races. Steelhead is the name given to members of O. mykiss that are anadromous, undertaking at least one sea migration prior to a return to freshwater to spawn, usually in their natal stream. Non-anadromous fish from the species are called rainbow or redband trout and are distributed throughout western north America. Over the past century, dam construction and habitat degradation have left native steelhead populations fragmented and greatly reduced, with many now protected by the US Endangered Species Act (ESA; NOAA 2006). Concurrently, billions of hatchery-bred steelhead and rainbow trout have been released into watersheds throughout the native range as mitigation and to enhance fisheries. The importance of local adaptation in salmonid fishes is well established (Taylor 1991; Adkison 1995; Martínez et al. in press) and such adaptation can be disrupted by dam construction or other habitat modifications. In addition, hatchery fish often have lower fitness than naturally spawning individuals (McLean et al. 2003; Araki et al. 2008; Caroffino et al. 2008). Thus, human activities have great potential to impact salmonid adaptation and life-history characteristics, including migratory behavior and the genetic relationships among populations (Eldridge and Naish 2007; Haugen et al. 2008; Williams et al. 2008), but the evolutionary response of specific salmonid populations to a given impact is difficult to predict (Carlson and Seamons 2008).
The availability of steelhead population samples collected more than 100 years ago from several coastal California watersheds, prior to dam construction and the extensive hatchery stocking of the twentieth century, allowed us to evaluate the historical genetic population structure and diversity of steelhead populations. In California, six Distinct Population Segments (DPSs), formerly designated as Evolutionarily Significant Units, of steelhead have been delineated on the basis of genetic, geographic and ecological variation: five coastal DPSs and one in the Central Valley (Busby et al. 1996). All but one of these are protected as threatened or endangered under the US ESA and recovery plans are currently being developed for the California coast steelhead DPSs. We used mitochondrial DNA (mtDNA) sequence data to provide a historical reference with which to compare patterns of genetic variation seen in modern O. mykiss populations. If human-induced fragmentation and hatchery stocking have not greatly affected these populations, then the genetic structure of historical and modern populations is expected to be similar. In doing so, we test the hypothesis that the population genetic relationships of steelhead currently spawning in coastal California streams are representative of the steelhead populations historically present in those streams. The results of this study will help to inform state and federal managers involved in recovery planning.
Materials and methods
Samples and DNA extraction
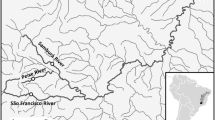
We obtained tissue samples of O. mykiss (identified as Salmo irideus or S. gairdneri) from 497 specimens housed at the Smithsonian Institution’s National Museum of Natural History that were originally collected from nine sites in six coastal basins in 1897 and 1909 (Table 1, Fig. 1; Snyder 1912). These specimens are currently stored in ethanol, although it is likely they were originally preserved in formalin. Small tissue samples (fin clips) were taken from the specimens and dried individually on blotter paper for transport to the laboratory. All subsequent historical sample handling was done in an isolated laboratory facility, occupied by the authors since construction, with new, dedicated pipettes and reagents. All other equipment was cleaned with bleach prior to use to avoid contamination with modern O. mykiss material. DNA extractions from historical samples were conducted manually using the DNeasy 96 tissue protocol (Qiagen, Inc.), with two negative extraction controls included in each 96-well extraction plate.
Map of coastal California watersheds from which O. mykiss were sampled for the present study. A single site was sampled in the South Fork Eel River (SFE), Coyote Creek (COY), and San Lorenzo River (SLO). Uvas (PJU) and Llagas (PJL) Creeks are tributaries of the Pajaro River, and the Arroyo Seco (SAS), San Antonio (SSA), and Nacimiento (SNC) Rivers are tributaries of the Salinas River. Grey area and lines in the inset map indicate current DPS delineations
For comparison, we analyzed 208 O. mykiss samples collected from approximately the same locations between 1997 and 2004 (Table 1). In addition, samples of seven rainbow trout strains from four hatcheries were analyzed: American River Hatchery, “Shasta” (SHT) and “Eagle” (EAG) strains; Hot Creek Hatchery, “Kamloops” (KAM) strain; Trinity River Hatchery steelhead (TRH); Fillmore Hatchery, “Virginia” (VIR), “Coleman”(COL), and “Mount Whitney” (WHT) strains. These hatchery strains represent all of the rainbow trout strains currently used for stocking, but do not necessarily include the wide variety of strains stocked into coastal California waters over the last century. DNA extraction from modern samples was conducted using the DNeasy 96 tissue protocol and processed on a BioRobot 3000 (Qiagen, Inc.) following standard laboratory procedures.
MtDNA amplification and sequencing
Primers that amplify approximately 193 base pairs of the right-domain of the mtDNA control region were used for all modern sample amplification and sequencing (P2 and S-Phe; Nielsen et al. 1994, 1998). However, due to degradation of DNA in the historical samples, amplification of the full P2/S-Phe region target was unsuccessful. We therefore designed primers (Table 2) to amplify two smaller, overlapping fragments that together cover 139 bp of the P2/S-Phe region, and include all but two of the sites known to be variable in O. mykiss (Graziano et al. 2005). We verified that all primer combinations amplify the same gene region by sequencing products from a single modern individual with all possible forward and reverse primer combinations and compiling the resulting sequences with complete P2/S-Phe sequences from other modern individuals to ensure correct alignment. All sequences were then trimmed so that only variable sites within the identical 139 bp region were considered in the analysis of all modern and historical samples.
All PCR amplifications included at least one negative control, and amplification of historical samples was conducted in an isolated laboratory using dedicated equipment as described above. PCRs were performed in 30 or 40 μl volumes with 4 μl DNA extract (undiluted for historical samples and diluted 1:20 with ddH20 for the modern samples). Each reaction contained 1× PCR buffer (Applied Biosystems, Inc.), 5.0 mM MgCl2, 0.1 mM each dNTP, 1 μM each primer, 1 μl bovine serum albumin buffer, 1 Unit DNA polymerase (AmpliTaq Gold, Applied Biosystems, Inc.), and 4 μl of DNA template. For historical samples that produced a weak initial amplification, a second identical PCR was performed using the first PCR product as template. Following PCR, amplification products were visually verified on 2% agarose gels stained with ethidium bromide, and successful amplifications were directly purified using PCR purification spin columns (Qiagen, Inc.) or excised from the agarose following electrophoresis and purified using the Qiaquick gel purification kit (Qiagen, Inc.). Sequencing reactions were carried out using BigDye cycle sequencing reagents (v1.1 and v3.1, Applied Biosystems, Inc.) and purified using Sephadex prior to electrophoresis on ABI 377 or 3730 automated sequencers. Sequences were verified and aligned using Sequencher (Gene Codes, Inc.). Sequencing of modern P2/S-Phe and historical Fragment1 products was done using the amplification primers. However, the short length of Fragment 2 made it difficult to sequence, so oligonucleotide extensions of standard M13 sequences were added to the 5′ ends of the Frag2F and Frag2R primers (Table 2). The resulting PCR products were 33 bp longer and were sequenced using complementary M13 sequencing primers.
Data analysis
All population genetic analyses were based on the observed frequencies of uniquely identifiable haplotype sequences. Pairwise FST estimates, exact tests for population differentiation, and AMOVA were done using ARLEQUIN 3.11 (Excoffier et al. 2005) with Bonferroni correction for multiple tests. Gene diversity and haplotype richness (based on rarefaction to the minimum sample size of n = 10) were calculated using CONTRIB (Petit et al. 1998). The relationships between genetic differentiation (represented by FST and [FST/(1 − FST)]) and several measures of the geographic distances among sampling locations for the historical and modern populations were evaluated to detect IBD with the method of Slatkin (1993) using the ISOLDE option in Genepop (Raymond and Rousset 1995). Geographic distances were estimated as the total combined river channel and coastal contour distances between sampling locations (i.e. river plus marine), as well as considering only marine distances among river mouths. Statistical significance based on Spearman’s rank correlation coefficient was evaluated using Mantel tests (Mantel 1967).
To assess the influence of sampling error on the estimation of IBD, we used a resampling approach to create 1,000 bootstrap samples from our data set by resampling with replacement and matching the actual sample sizes for each population. The pairwise FST values between all populations were calculated from each bootstrap replicate and used to assess the strength of IBD. The distribution of bootstrap-generated slopes of IBD correlations for the historical and modern samples then provides an indication of the effect of sampling error (i.e. small sample sizes) on the observed comparison of IBD between the historical and modern samples.
Results
Genetic data
Initial tests showed variability among samples sites in the quality of DNA recovered, and many individuals failed to produce usable PCR products with any primer combination. Complete Frag1 + Frag2 sequences were obtained from a total of 180 historical specimens from eight O. mykiss populations in five central California rivers (Table 1). An additional 25 complete or partial sequences were obtained from specimens sampled from Lagunitas Creek (a small creek approx. 50 km north of San Francisco, CA). However, examination of these sequences revealed that almost all of them were in fact from coho salmon (Oncorhynchus kisutch), a sympatric congener of O. mykiss, and were apparently misidentified in the original collection. Thus, these sequences were excluded from further consideration in the present analysis.
Graziano et al. (2005) standardized the nomenclature for 29 mtDNA control region haplotypes previously observed in O. mykiss based on the P2/S-Phe region. However, the Frag1 + Frag2 alignment, which covered nucleotide sites 1021–1109 (Graziano et al. 2005), cannot distinguish several known haplotypes described by Graziano et al. (2005), which are defined by these and nine additional variable nucleotide sites in the full P2/S-Phe region. In addition, we observed four new haplotypes, two in historical specimens and two in modern individuals (Pearse and Garza 2008). In total, eleven unique haplotype sequences were observed based on the shorter Frag 1/2 sequences (Table 3) and all population genetic analyses were based on the frequencies of these identifiable haplotypes. All unique sequences have been deposited in GenBank (Accession numbers HQ682186–HQ682189).
Genetic diversity and differentiation
Six haplotypes were seen in both modern and historical collections, two were seen only in historical individuals, one was seen in a single modern individual, and two haplotypes were seen only in hatchery rainbow trout. Although no site had evidence for complete haplotype replacement, in 10 cases a haplotype that was historically present at a given site was not observed in the corresponding modern sample (Table 3). Conversely, there were seven instances in which a haplotype was observed in a modern population but was not observed in the historical sample from the same location. For example, in the PJL population, haplotypes A, E, and G were found in the historical sample, but haplotypes B, C, and E were observed in the modern sample (Table 3). On average, the historical population samples had a larger absolute number of haplotypes and greater haplotype richness (HR) per population than did the modern populations (Table 3), although the differences were not statistically significant (3.63 vs. 3.25 and 2.20 vs. 1.92, respectively, paired t tests, n.s.). However, overall number and per population HR were significantly lower in the seven hatchery strains (1.86 and 0.77, respectively, ANOVA, P < 0.001) than in either the historical or modern natural populations (Table 3).
Half of the natural populations had historical haplotype frequencies that were significantly different from the corresponding modern sample (exact tests for population differentiation, α < 0.05), and pairwise FST values for such temporal comparisons of the same location ranged from −0.041 to 0.501 (Table 4). Finally, in order to test for a relationship between genetic differentiation in the historical and modern samples, we calculated the mean pairwise genetic differentiation of each historical and modern sample against its contemporary populations. We found no correlation between either mean pairwise FST (R2 = 0.013, n.s.), or the allelic richness (HR) of historical versus modern comparisons (R2 = 0.043, n.s.), suggesting that substantial change has occurred in many of the populations over time.
Isolation-by-distance
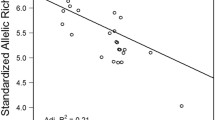
We found no significant relationship between genetic distance, as measured by pairwise FST values, and geographic distances between river sampling sites in the modern populations (R2 = 0.07, n.s.; Fig. 2a). In contrast, a strong and highly significant relationship was observed for the same parameters in the historical populations (R2 = 0.72, P < 0.01; Fig. 2b), suggesting that less than 10% of the historical level of IBD remains in the modern populations. This relationship remained highly significant regardless of the combination of FST or [FST/(1 − FST)] with geographic distance or the natural log of geographic distance employed (Rousset 1997). To evaluate the sensitivity of the IBD results to different estimators of genetic and geographic distance, we also estimated genetic distances by pooling the haplotype frequencies for all individuals within the Pajaro and Salinas River watersheds, respectively, and comparing these with geographic distances considering only the coastal marine distances among river-mouths. Importantly, this analysis accounts for the uncertainty in the exact location from which some of the historical collections were taken (Snyder 1912). In all cases, historical IBD was strong and significant while modern IBD was not significantly different from zero (mean historical R2 = 0.78, range 0.72–0.89; mean modern R2 = 0.06, range 0.005–0.11). Thus, these results are robust to the use of diverse estimates of genetic and geographic distance and indicate that IBD was historically much stronger than it is today.
To further strengthen our inference of significantly greater IBD in the historical samples compared with the modern populations, and to evaluate the possibility that sampling variance might create a spurious pattern, we used bootstrap resampling to test the hypothesis that sample size alone could account for the high observed IBD in the historical samples or the lack of IBD detected in the modern samples. We consistently detected a significant signal of greater isolation-by-distance in the historical samples, and conversely a lack of isolation-by-distance in the modern samples, despite bootstrapping over the small sample sizes from some populations. Compared with the observed difference of 65% between the slopes of IBD in the historical and modern populations, our simulated samples detected an equal or greater difference in 75% of the cases, and a difference of 46% or more in 95% of the simulations. Therefore, if the observed difference between the historical and modern samples were due to small sample size, it is unlikely that it would persist in bootstrapped samples.
The loss of IBD observed in modern populations is influenced by changes in genetic distance between both neighboring and distant population pairs (Table 4; Fig. 2a, b). Among the cluster of streams tributary to Monterey Bay (SLO, PJU, PJL, SAS, SSA, SNC), genetic differentiation has more than doubled (mean pairwise FST: historical = 0.11, modern = 0.25; paired t test, P < 0.05). Conversely, average genetic differentiation between the Monterey Bay populations and the two more distant populations (Eel River, SFE, and Coyote Creek, COY) has been significantly reduced (mean pairwise FST: historical = 0.39, modern = 0.26; paired t test, P < 0.05). Analysis of molecular variance (AMOVA) is consistent with the IBD and pairwise F ST results; between-group differentiation for the grouping Monterey Bay versus Coyote Creek versus Eel River historically explained 28.8% of the observed genetic variation, but today explains less than 1% (Table 5). This change is accompanied by an increase from 8 to 26% in the percent of variation explained by differentiation among the Monterey Bay tributary populations, which are all <30 marine km apart (Table 5). Thus, changes over the past century have both increased divergence among proximate populations and eroded differences between distant populations, such that patterns of genetic diversity in modern steelhead no longer reflect their historical patterns of migration.
Discussion
Studies of the genetic structure of extant natural populations of many taxa frequently find a significant signal of isolation-by-distance (e.g. salmonids (Palstra et al. 2007), including California steelhead (Garza et al. 2004; Pearse et al. 2007), cod (Pogson et al. 2001), oysters (Peterson and Denno 1998), insects (Rose et al. 2006), and plants (Cruse-Sanders and Hamrick 2004), but see Pearse et al. (2006) for an exception in turtles). However, without a historical reference point, there is no way to know if the populations are in migration-drift equilibrium or representative of their natural state. In the case of coastal California steelhead, data from 18 microsatellite loci in 60 modern populations documented moderate IBD (R2 = 0.193, P < 0.01; R2 = 0.191, n.s. when only the eight populations studied here are considered; Garza et al. 2004; Clemento et al. 2009). While these nuclear data suggest a slightly stronger correlation between geographic and genetic distance than do our modern mtDNA sequences, they are consistent with the hypothesis that the historically-strong IBD we document here has been greatly reduced over the past hundred years.
The present study of historical population genetic structure of coastal California steelhead found that some populations displayed dramatic shifts in haplotype frequency between the historical and modern collections, whereas others remained almost unchanged (Table 3). Although measures of average within-population genetic diversity were lower in modern populations, the observed differences were not significant or consistent across all population pairs (Table 3). However, the strong correlation we observed in the historical mtDNA data between genetic divergence and the geographic distance separating each pair of populations indicates that a natural pattern of genetic differentiation proportional to geographic distance has been virtually erased in ESA-listed coastal steelhead, presumably due to human impacts over the past century. To the extent that these populations were historically adapted to the local ecological conditions of these watersheds, the disruption of equilibrium between geographic and genetic distance may also have affected such adaptation.
Although we cannot directly determine which factors are most responsible for the observed changes in steelhead population genetic structure, the most likely impacts appear to be fragmentation due to dam construction and habitat degradation, and genetic introgression by conspecific, but non-local hatchery rainbow trout (Hindar et al. 1991). The rivers sampled in the present study have been heavily impacted by human modifications over the last century. For example, the Salinas River supports an extensive agricultural industry, and the lower mainstem runs completely dry during many summer months, leaving fish isolated in tributary waters. Dams on the San Antonio and Nacimiento Rivers further disrupt migration to these tributary streams, and dams and other diversion structures also affect migration on Coyote Creek and the Pajaro River tributaries (Uvas and Llagas Creeks). Microsatellite data indicate that southern California O. mykiss populations above dams are primarily descended from steelhead trapped at construction and not from the hatchery rainbow trout subsequently released into the dam reservoirs, although some introgression has occurred (Clemento et al. 2009). However, other studies that have compared pre- and post-stocking genetic composition of salmonid populations have shown a range of genetic changes following stocking (e.g. brown trout, Hansen 2002; Hansen et al. 2009; coho salmon, Eldridge and Naish 2007; grayling, Koskinen et al. 2002; Susnik et al. 2004). Thus, care should be taken when using modern data from human-impacted populations to infer historical genetic patterns.
The present results demonstrate the historical importance of migration in the maintenance of equilibrium, and therefore effective population size, of coastal steelhead, including populations up to 200 km from the ocean. The demonstration of the importance of migration has direct application to management and conservation of these ESA-protected fish populations, as well as more general habitat conservation plans and the design of coastal marine reserves (Carr et al. 2003). The direct evaluation of historical population structure of steelhead also provides a compelling example of the value of natural history museum collections in providing a historical baseline for comparison with modern populations and planning for their continued existence (Nielsen and Hansen 2008).
References
Adkison MA (1995) Population differentiation in Pacific salmons: local adaptation genetic drift, or the environment? Can J Fish Aquat Sci 52:2762–2777
Araki H, Berejikian BA, Ford MJ, Blouin MS (2008) Fitness of hatchery-reared salmonids in the wild. Evol Appl 1:342–355
Busby PJ, Wainwright TC, Bryant GJ, Lierheimer LJ, Waples RS, Waknitz FW, Lagomarsino IV (1996) Status Review of West Coast steelhead from Washington, Idaho, Oregon, and California. NOAA Technical Memorandum, NMFS-NWFSC-27
Carlson SM, Seamons TR (2008) A review of quantitative genetic components of fitness in salmonids: implications for adaptation to future change. Evol Appl 1:222–238
Caroffino DC, Miller LM, Kapuscinski AR, Ostazeski JJ (2008) Stocking success of local-origin fry and impact of hatchery ancestry: monitoring a new steelhead (Oncorhynchus mykiss) stocking program in a Minnesota tributary to Lake Superior. Can J Fish Aquat Sci 65:309–318
Carr MH, Neigel JE, Estes JA, Andelman S, Warner RR, Largier JL (2003) Comparing marine and terrestrial ecosystems: implications for the design of coastal marine reserves. Ecol Appl 13:S90–S107
Clemento A, Anderson EC, Boughton D, Girman D, Garza JC (2009) Population genetic structure and ancestry of Oncorhynchus mykiss populations above and below dams in south-central California. Conserv Genet 10:1321–1336
Crandall KA, Bininda-Edmonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol Biol 15:290–295
Cruse-Sanders JM, Hamrick JL (2004) Genetic diversity in harvested and protected populations of wild American ginseng, Panax quinquefolius L. (Araliaceae). Am J Bot 91:540–548
Eldridge WH, Naish KA (2007) Long-term effects of translocation and release numbers on fine-scale population structure among coho salmon (Oncorhynchus kisutch). Mol Ecol 16:2407–2421
Excoffier L, Laval G, Schneider S (2005) ARLEQUIN ver. 3.0: an integrated software package for population genetic data analysis. Evol Bioinform Online 1:47–50
Garza JC, Gilbert-Horvath E, Spence BC, Williams TH, Fish H, Gough S, Anderson JH, Hamm D (2004) Population structure and history of steelhead trout in California. In: Irvine J et al (eds) Workshop on application of stock identification in defining marine distribution and migration of salmon. North Pacific Anadromous Fish Commission Technical Report 5, pp 129–131
Graziano SL, Brown KH, Nielsen JL (2005) Nomenclature of mitochondrial DNA haplotypes for Oncorhynchus mykiss. Trans Am Fish Soc 134:1271–1273
Hansen MM (2002) Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Mol Ecol 11:1003–1015
Hansen MM, Fraser DJ, Meier K, Mensberg K-LD (2009) Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Mol Ecol 18:2549–2562
Haugen TO, Aass P, Stenseth NC, VØllestad LA (2008) Changes in selection and evolutionary responses in migratory brown trout following the construction of a fish ladder. Evol Appl 1:319–335
Heath DD, Busch C, Kelly J, Atagi DY (2002) Temporal change in genetic structure and effective population size in steelhead trout (Oncorhynchus mykiss). Mol Ecol 11:197–214
Hindar K, Ryman N, Utter F (1991) Genetic effects of cultured fish on natural fish populations. Can J Fish Aquat Sci 48:945–957
Jackson JBC, Kirby MX, Berger WH, Bjorndal KA, Botsford LW, Bourque BJ, Bradbury RH, Cooke R, Erlandson J, Estes JA, Hughes TP, Kidwell S, Lange CB, Lenihan HS, Pandolfi JM, Peterson CH, Steneck RS, Tegner MJ, Warner RR (2001) Historical overfishing and the recent collapse of coastal ecosystems. Science 293:629–638
Koskinen MT, Sundell P, Piironen J, Primmer CR (2002) Genetic assessment of spatiotemporal evolutionary relationships and stocking effects in grayling (Thymallus thymallus, Salmonidae). Ecol Lett 5:193–205
Leonard JA (2008) Ancient DNA applications for wildlife conservation. Mol Ecol 17:4186–4196
Leonard JA, Vila C, Wayne RK (2005) Legacy lost: genetic variability and population size of extirpated US grey wolves (Canis lupus). Mol Ecol 14:9–17
Lozier JD, Cameron SA (2009) Comparative genetic analysis of historical and contemporary collections highlight contrasting demographic histories for the bumble bees Bombus pensylvanicus and B. impatiens in Illinois. Mol Ecol 18:1875–1886
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Martínez A, Garza JC, Pearse DE (in press) A microsatellite genome screen identifies chromosomal regions under differential selection in steelhead and rainbow trout (Oncorhynchus mykiss). Trans Am Fish Soc
McLean JE, Bentzen P, Quinn TP (2003) Differential reproductive success of sympatric, naturally spawning hatchery and wild steelhead trout (Oncorhynchus mykiss) through the adult stage. Can J Fish Aquat Sci 60:433–440
National Oceanic and Atmospheric Administration (2006) Endangered and threatened species: final listing determinations for 10 distinct population segments of west coast steelhead. Fed Regist 71:834–862
Newsome SD, Etnier MA, Gifford-Gonzalez D, Phillips DL, van Tuinen M, Hadly EA, Costa DP, Kennett DJ, Guilderson TP, Koch PL (2007) The shifting baseline of northern fur seal ecology in the northeast Pacific Ocean. Proc Natl Acad Sci USA 104:9709–9714
Nielsen EE, Hansen MM (2008) Waking the dead: the value of population genetic analyses of historical samples. Fish Fish 7:450–461
Nielsen JL, Gan C, Thomas WK (1994) Differences in genetic diversity for mitochondrial DNA between hatchery and wild populations of Oncorhynchus. Can J Fish Aquat Sci 51:290–297
Nielsen JL, Fountain MC, Favela JC, Cobble K, Jensen BL (1998) Oncorhynchus at the southern extent of their range: a study of mtDNA control-region sequence with special reference to an undescribed subspecies of O. mykiss from Mexico. Environ Biol Fish 51:7–23
Palstra FP, O’Connell MF, Ruzzante DE (2007) Population structure and gene flow reversals in Atlantic salmon (Salmo salar) over contemporary and long-term temporal scales: effects of population size and life history. Mol Ecol 16:4504–4522
Pauly D (1995) Anecdotes and the shifting baseline syndrome of fisheries. Trends Ecol Evol 10:430
Pearse DE, Garza JC (2008) Historical baseline for genetic monitoring of coastal California steelhead, Oncorhynchus mykiss. California Department of Fish and Game Fisheries Restoration Grant Program Report #0510530
Pearse DE, Arndt AD, Valenzuela N, Miller BA, Cantarelli V, Sites JW Jr (2006) Estimating population structure under non-equilibrium conditions in a conservation context: Continent-wide population genetics of the giant Amazon River turtle Podocnemis expansa (Chelonia; Podocnemidae). Mol Ecol 15:985–1006
Pearse DE, Donohoe CJ, Garza JC (2007) Population genetics of steelhead (Oncorhynchus mykiss) in the Klamath River. Environ Biol Fish 80:377–387
Peterson MA, Denno RF (1998) The influence of dispersal and diet breadth on patterns of genetic isolation by distance in phytophagous insects. Am Nat 152:428–446
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pinnegar JK, Engelhard GH (2008) The ‘shifting baseline’ phenomenon: a global perspective. Rev Fish Biol Fish 18:1–16
Pogson GH, Taggart CT, Mesa KA, Boutilier RG (2001) Isolation by distance in the Atlantic cod, Gadus morhua, at large and small geographic scales. Evolution 55:131–146
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rose CG, Paynter KT, Hare MP (2006) Isolation by distance in the eastern oyster, Crassostrea virginica, in Chesapeake Bay. J Hered 97:158–170
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Snyder JO (1912) The fishes of the streams tributary to Monterey Bay, California. Bull Bur Fish 32:49–72
Susnik S, Berrebi P, Dovc P, Hansen MM, Snoj A (2004) Genetic introgression between wild and stocked salmonids and the prospects for using molecular markers in population rehabilitation: the case of the Adriatic grayling (Thymallus thymallus L. 1785). Heredity 93:273–282
Taylor EB (1991) A review of local adaptation in Salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture 98:185–207
Wandeler P, Hoeck PEA, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22:634–642
Williams JG, Zabel RW, Waples RS, Hutchings JA, Connor WP (2008) Potential for anthropogenic disturbances to influence evolutionary change in the life history of a threatened salmonid. Evol Appl 1:271–285
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
This study was partially funded by California Department of Fish and Game Fisheries Restoration Grant P0510530 to DEP and JCG. We thank A. Abadia, A. Clemento, M. Jones, and A. Martinez for assistance, and E. C. Anderson for conducting the bootstrap resampling analysis. J. Finan (National Museum of Natural History) assisted with sampling of the historical specimens. D. Boughton, H. Fish, M. Moore, L. Porcella, D. Salsbery, B. Spence, and T. Williams contributed to the collection of modern samples.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Pearse, D.E., Martinez, E. & Garza, J.C. Disruption of historical patterns of isolation by distance in coastal steelhead. Conserv Genet 12, 691–700 (2011). https://doi.org/10.1007/s10592-010-0175-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0175-8