Abstract
Ackee (Blighia sapida) is a native multipurpose species important for the livelihoods of the rural populations in Benin. Trees are found in natural forests or are managed by farmers in different traditional agroforestry systems. Genetic variation at amplified fragment length polymorphism (AFLP) markers, four nuclear microsatellites (nSSRs) and one chloroplast microsatellite (cpSSR) were investigated in 279 individuals from six wild and eight cultivated populations from Benin. The AFLP data revealed moderate levels of diversity of ackee in Benin (mean diversity values are proportion of polymorphic loci = 52.8% and Nei’s gene diversity = 0.157, for 375 AFLP fragments). The mean diversity values based on nSSR-markers are expected heterozygosity = 0.286, allelic richness = 2.77. Genetic variation of wild and cultivated populations did not differ markedly. AMOVA revealed that only 7.3 and 5.2% of the variation was partitioned among populations for nSSR- and AFLP-markers, respectively. A Mantel test based on these both marker-types revealed significant correlations between population pairwise geographic distance and genetic differentiation. Differentiation among cultivated populations was higher than among wild populations. The only polymorphic chloroplast microsatellite marker (ccmp7) showed three haplotypes. Cultivated populations from northeastern Benin were fixed on one haplotype which was not observed elsewhere indicating a different origin of these populations possibly from neighboring Nigeria. Farmer-led domestication had an impact on the spatial distribution of genetic variation but did not result in significant losses of diversity within populations. Measures to conserve genetic resources of ackee in each of the three main bioclimatic zones in Benin are proposed.
Similar content being viewed by others
Introduction
Domestication is generally considered to be the end-point of a continuum that starts with exploitation of wild plants, continues through cultivation of plants selected from the wild but not yet genetically different from their wild progenitors, and terminates in fixation, through human selection, of morphological and hence genetic differences distinguishing a domesticated variety from its wild relatives (Pickersgill 2007). The phenotypic changes associated with adaptation under domestication are commonly referred to as the domestication syndrome. Common features of the domestication syndrome are gigantism (especially of the harvested part of the plant), reduced branching, loss or reduction of seed dispersal, loss of seed dormancy, changes in photoperiod sensitivity, and loss or reduction of toxic compounds (Gepts 2004). These changes depended upon the degree of domestication and on life history traits of the species in question. Most vegetatively propagated root crops and perennial fruit crops like Blighia sapida show fewer features of the domestication syndrome than annual seed crops; and domestication may occur more slowly because fewer sexual generations occur in a given period of time (Pickersgill 2007). Clement (1999) proposed two intermediate categories, ‘incipiently domesticated’ and ‘semi-domesticated’, to cover the spectrum of changes resulting from human interactions with species of fruit trees.
Blighia sapida is a woody perennial multipurpose fruit tree species native to the Guinean forests of West Africa. The fleshy arils of the ripened fruits are edible. Seeds and capsules of the fruits are used for soap-making and for fishing, and all parts of the tree have medicinal properties. Fresh arils, dried arils and soap are traded in local and regional markets in Benin providing substantial revenues for farmers, especially women (Dossou et al. 2004; Ekué et al. 2004). Although overlooked by researchers in the region, the tree is highly valued by farmers and is an important component of traditional agroforestry systems in Benin. Recently, ackee has emerged as high-priority species for domestication in Benin after a national survey and ranking of Non-Timber Forest Products (NTFPs) (Codjia et al. 2003). B. sapida is widely cultivated in Jamaica where it had been introduced by slave traders during the eighteenth century (ICRAF 2009) with an annual turnover of approximately US $ 400 million in 2005 for the trade of the arils of the fruits (Pen 2006).
In Benin previous studies based on farmers’ perceptions/folk classification and morphological characterization showed that there is variability within the species across different climatic conditions. Farmers use their own criteria (bark color, tree height, tree diameter, fruits productivity, fruits size, phenology etc.) to differentiate between trees from the Sudanian and Guineo-Sudanian zones in the North compared to the Guinean or Guineo-Congolian zone in the South. For example, trees are on average taller and fruits are bigger in the North than in the South. People have reported nine criteria mostly related to the fruits to differentiate between different types of ackee. Ackee phenotypes with preferred fruit traits (size, color, aril taste and oil content) are perceived by local people to be more abundant in ‘cultivated’ populations (field/fallow stands) than in unmanaged ‘wild’ populations (forest stands) (Ekué et al. 2004, 2009b, 2010). Thus, B. sapida can be considered as either ‘incipiently domesticated’ or ‘semi-domesticated’ in traditional agroforestry systems in Benin, and therefore provides an excellent opportunity to document the effect of ongoing domestication on levels of genetic variation in cultivated populations.
In this study we used Amplified Fragment Length Polymorphism (AFLP) markers, nuclear microsatellites markers recently successfully transferred from Litchi chinensis to B. sapida (Ekué et al. 2009a) and one chloroplast microsatellite marker to investigate genetic variation patterns in natural and managed in situ and cultivated populations of ackee in Benin. AFLP (Vos et al. 1995) markers cover the whole genome, and have proven to be useful in assessing genetic variation and taxonomic relatedness in a large variety of wild and cultivated plant species (Miller and Schaal 2006; Emshwiller et al. 2009). Nuclear microsatellites (nSSRs) are expected to be particularity effective to test for genetic bottlenecks associated with domestication (Kelly et al. 2004; Hollingsworth et al. 2005). Cross-species amplification of the Litchi chinensis primers used was previously reported in Euphoria longan and Nephelium rambutan-ake; all four species belong to the Sapindaceae family (Viruel and Hormaza 2004; Sim et al. 2005). Chloroplast microsatellite markers are frequently used in phylogeographic analyses of plant species (Fontaine et al. 2004; Muller et al. 2009) but also to infer the origin of planted tree stands on-farm (Dawson et al. 2008).
Our mains objectives were (i) to characterize the level and organization of genetic diversity within populations, (ii) to assess the impact of human management practices on the genetic diversity and structure and (iii) to infer the origin of cultivated populations in Benin.
Materials and methods
Plant material and DNA isolation
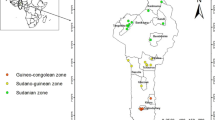
Fourteen populations covering the full range of the species in Benin, situated in different climatic zones and having different management status (‘wild’ or ‘cultivated’) were sampled. The wild populations are composed of ackee trees that have grown up naturally; they have no level of management and there is no regeneration practice put in place by local people. In contrast, the cultivated populations are composed mainly of ackee trees planted by farmers with sometimes only few remnants of natural trees; those populations have moderate to high levels of management designed to improve the production including regeneration practices for the best producing trees (Ekué et al. 2009b, 2010). Within each location, adult trees were randomly sampled. A minimum distance of 100 m between individual trees was respected to avoid selection of closely related neighboring individuals. Only adult trees that have already fruited once were considered in this study. In total, leaves were collected from 279 individual trees and dried in the field using silica gel. Table 1 describes the geographic location of each population, the sample size and the status. Six populations were wild and eight populations were cultivated (Fig. 1).
Total genomic DNA was extracted from a small slice (approximately 1 cm2) of dried leaf tissue following the DNeasy® Plant Kit protocol of the manufacturer (Qiagen, Hilden, Germany) and adding polyvinylphenol (PVP) to the AP1 lysis buffer up to 2.6%.
AFLP analysis
The AFLP procedure followed the protocol of Vos et al. (1995) with minor modifications. Genomic DNA (4 μl) was digested simultaneously with the two restriction enzymes MseI and EcoRI. Double-stranded MseI and EcoRI adaptors were ligated to the ends of the restriction fragments to generate template DNA for polymerase chain reaction (PCR) amplification. The restriction/ligation was performed overnight at room temperature. Preamplification reactions were carried out with diluted DNA from the ligation reaction and the preselective primer pairs E01/M01 each having the selective nucleotide A and G, respectively. The reactions were programmed to start at 72°C for 2 min, 20 cycles each consisting at 94°C for 10 s, at 56°C for 30 s, at 72°C for 2 min, and then at 60°C for 30 min. Selective amplifications were carried out using diluted DNA from the preamplification reaction and the primer combination MseI-AGA and EcoRI-GAT. Primer MseI-AGA was labeled with the fluorescent dye HEX. The thermal cycler was programmed to start at 94°C for 2 min, 10 cycles, each consisting of 10 s at 94°C, 30 s at 65°C and 2 min at 72°C. The 65°C annealing temperature of the first cycle is subsequently reduced by 1°C for the next 10 cycles and continued at 56°C for 30 s for the remaining 23 cycles, and ends with a final extension step at 60°C for 30 min. This primer combination was adopted from a test of six primer combinations in eight samples of B. sapida and selected based on the production of reproducible, clear, easily and unambiguously scorable fragments. All PCR reactions were conducted in a Peltier Thermal Cycler (PTC-200 version 4.0, MJ Research). The amplified restriction products were resolved electrophoretically on an ABI Genetic Analyzer 3100 together with the internal size standard GeneScan 500 ROX (fluorescent dye ROX) from Applied Biosystems. The size of the AFLP fragments was determined with the software packages GeneScan 3.7 and Genotyper 3.7 (Applied Biosystems Inc.).
Nuclear microsatellites
Four polymorphic primers pairs (LMLY4, LMLY5, LMLY6, LMLY7) transferred from Litchi chinensis (Ekué et al. 2009b) were investigated. PCR amplifications were conducted in a 10 μl volume containing 1 μl of genomic DNA (about 10 ng), 5 μl of HotStarTaq Master Mix (0.1 U/μl Taq DNA polymerase, 3 mM MgCl2, 400 μM dNTPs, Qiagen, Hilden, Germany) and 5 pmol of each primer. The PCR protocol consisted of an initial denaturation step of 95°C for 15 min, followed by 35 cycles of 94°C for 30 s (denaturation), 50°C for 1 min (annealing), 72°C for 1 min (extension) and a final extension step of 72°C for 10 min. For primers LMLY4 and LMLY6, the annealing temperature was 45 and 39.4°C, respectively; and for both primers, the duration of the denaturation and annealing during the 35 cycles was 1 min. The technical equipments used for PCR reactions and visualization of amplified products, and the software used for fragments size determination were the same as mentioned above for AFLPs.
Chloroplast microsatellites and sequencing
A total of ten universal microsatellite primer pairs (ccmp1 to ccmp10) described by Weising and Gardner (1999) and four primer pairs (udt1, udt4, ucd4, ukk4) described by Deguilloux et al. (2003) were tested within a subset of 12 samples from all populations. The amplification reaction was performed in a 15 μl volume containing 5.1 μl distilled water (Qiagen, Hilden), 7.5 μl of HotStarTaq (0.735 units) Master Mix (Qiagen, Hilden), 1.2 μl primer (5 pmol/μl) and 1.2 μl of genomic DNA (about 10 ng). The reaction profile was: 95°C for 15 min; 35 cycles for 1 min at 94°C, 1 min at 50°C, 1 min at 72°C and a final extension step for 10 min at 72°C. PCR reactions were conducted in a Peltier Thermal Cycler (PTC-200 version 4.0, MJ Research). The resolution of the amplified products and the determination of fragments sizes followed the same methods described above for AFLPs.
Comparative sequencing of 18 fragments derived from the primer pair ccmp7 was performed to confirm the amplified regions and to determine the molecular basis for size variation. The amplification products were purified using the QIAquick Gel Extraction kit (Qiagen®, Germany) following the manufacturer’s specifications. The purified PCR products were cloned in a pCR2.1 vector using a TOPO TA cloning® kit (Invitrogen, Carlsbad, CA). The inserts were amplified by colony PCR using M13 forward (−20) (5′-GTAAAACGACGGCCAG-3′) and M13 reverse (5′-CAGGAAACAGCTATGAC-3′) primers, visualized by agarose gel electrophoresis, excised from the gel and purified. Three to six different clones of the fragments were sequenced using both M13 forward and M13 reverse primers in order to control for Taq polymerase errors.
The sequencing reactions were carried out with the Big Dye® Terminator v.3.1. Cycle Sequencing Kit (Applied Biosystems) based on the dideoxy-mediated chain termination method (Sanger et al. 1977). Sequencing reactions were run on the ABI 3100 Genetic Analyser (Applied Biosystems). The sequence data have been stored in the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/) with the accession numbers FN543506, FN543507 and FN543508.
Data analysis
Diversity analyses
For AFLPs, the presence or absence of amplification products was scored for each sample within a readable range of 75–500 bp and assembled as a binary (1/0) matrix. Electropherograms were carefully checked individually to avoid possible misinterpretations because of automated fragment scoring. AFLP data were analyzed first using AFLPDiv version 1.1 (available at http://www.pierroton.inra.fr/genetics/labo/Software/Aflpdiv; Coart et al. 2005) to compute the percentage of polymorphic loci (PLP) and the number of phenotypes Br (or ‘band richness’) expected at each ‘locus’ (=scored AFLP fragment) in each population after rarefaction to the smallest sample size (12 in this case) (see El Mousadik and Petit 1996). A locus is considered polymorphic at the 1% level if 1.01 < Br < 1.99. Non-random association between markers (linkage disequilibrium) was tested for all marker pairs in each population as implemented in ARLEQUIN version 3.11 (Excoffier et al. 2007). AFLP-SURV 1.0 (Vekemans 2002) was used to estimate allelic frequencies at AFLP loci from the observed fragment frequencies using the Bayesian approach proposed by Zhivotovsky (1999) for diploid species. A non-uniform prior distribution of allelic frequencies was assumed with its parameters derived from the observed distribution of fragment frequencies among loci (see note 4 in Zhivotovsky (1999). Allele frequencies were then used to calculate genetic differentiation between populations (F ST) and expected heterozygosity or Nei’s gene diversity (H j) (Lynch and Milligan 1994). Theses F ST and H j estimates were calculated for different values of F IS (0, 0.05 and 0.08), as no data about the level of inbreeding within the populations were available. Comparative assessments of mean and total genetic diversity parameters (H j, PPL and Br) were conducted between population categories (wild vs. cultivated), and the significance was tested by a t-test for independent samples using STATISTICA (version 7.0, 2004, StatSoft, Inc., Tulsa, OK).
For nuclear microsatellites (nSSRs), the number of alleles (nanuc) and the unbiased estimates of observed (H O) and expected (H E) heterozygosities were calculated for each locus and averaged for each population over all loci using GDA 1.1 (Lewis and Zaykin 2001). Alleles were deemed private (A P) if they showed a frequency of more than 5% in one population and did not occur in any other population. To check if diversity estimates were affected by differences in sample sizes, we calculated the allelic richness A R as a standardized measure of the number of alleles per locus corrected by the sample size using FSTAT v2.9.3.2 (Goudet 2001). Finally, a comparative assessment of A R and H E between wild and cultivated populations was undertaken with FSTAT v2.9.3.2. A two-tailed approach was used to compare variation among populations within categories to that detected between categories, and 1000 random permutations of the data were performed, to test for significance (Goudet 2001).
Since only one polymorphic locus was observed, a chloroplast haplotype was defined by the corresponding allele size at the locus ccmp7. The frequency of each haplotype at the locus ccmp7 was calculated per population and management category.
Population differentiation
For AFLPs, genetic distances between pairs of populations were calculated according to Nei (1978). To examine the genetic relationship among populations, a cluster diagram was generated from distance values using the Unweighted Pair-Group Method of Arithmetic averages (UPGMA) modified from the Neighbor procedure of PHYLIP version 3.5 (Felsenstein 1993).
For nSSRs, the pairwise differentiation between populations (F STnuc) was assessed by the θ-statistics of Weir and Cockerham (1984 ). The significance of pairwise population differentiation was tested using the log-likelihood statistics G (Goudet et al. 1996) implemented in FSTAT (v2.9.3.2). Significance values were calculated after sequential Bonferroni correction (Rice 1989). The matrix of genetic distances was calculated using the Cavalli-Sforza distance D2 (Cavalli-Sforza and Edwards 1967), which is expected to describe the structure of populations according to their evolutionary history. This matrix was used to perform a cluster analysis using the neighbor-joining method and Populations 1.2.30 software (Langella 1999).
For AFLP and nSSR markers, Mantel tests (Mantel 1967) were used to test the correlation between F ST/(1 − F ST) and the natural logarithm of geographic distance between sampled sites using GenAlEx 6.2 (Peakall and Smouse 2006): the two-dimensional model was used as suggested by Rousset (1996) because populations were distributed on a non-linear range. The Mantel test was performed also separately for the wild populations alone. An analysis of molecular variance (AMOVA; Excoffier et al. 1992) was carried out using the program ARLEQUIN 3.11 (Excoffier et al. 2007) to apportion variance applying the estimator of Weir and Cockerham (1984).
To test for genetic differentiation among population categories with AFLP and nSSR markers, F ST values were generated by population category and 10,000 permutations were used to test for significance using ARLEQUIN 3.11 (Excoffier et al. 2007). For cpSSR markers, the genetic differentiation (G ST) values were computed for the overall populations as well as for each management type using RAREFAC (Petit et al. 1998).
Sequence analysis
The sequences were aligned by using Codon Code Aligner (CodonCode cooperation, www.condoncode.com) and BioEdit version 7.0.0 (Hall 1999) using ClustalW multiple alignment (Thompson et al. 1994).
Correlation between AFLP and SSR markers
In order to test the consistency of SSR and AFLP markers in estimating genetic differentiation parameters, Mantel test was performed between pairwise F ST/(1 − F ST) estimates on the overall data set. A non parametric t-test of paired pairwise F ST values of the two markers was also performed (Woodhead et al. 2005).
Results
Genetic diversity and structure at AFLP markers
A total of 375 unambiguously scorable AFLP fragments, of which 86.9% were polymorphic in the total data set, were observed. Diversity statistics are summarized in Table 2. On average, nearly 53% of AFLP markers were polymorphic within populations. AFLP-SURV-derived genetic differentiation measures were: \( \mathop F\nolimits_{ST}^{{F_{IS} = 0}} = 0.034 \pm 0.245 \); \( \mathop F\nolimits_{ST}^{{F_{IS} = 0.05}} = 0.032 \pm 0.251 \) and \( \mathop F\nolimits_{ST}^{{F_{IS} = 0.08}} = 0.033 \pm 0.252 \). All F ST values were significantly different from zero (P < 0.01). Gene diversities (H j) for the three different F IS values were highly correlated (>99%). Thus, we used H j values for F IS = 0 for further calculations. The mean band richness [Br (12)] and Nei’s gene diversity (H j) were 1.452 and 0.157, respectively.
The test for non-random association between markers gave a mean percentage for associated loci of 13.4% (Table 2).
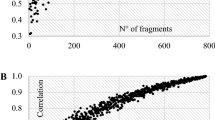
The analysis of molecular variance (AMOVA) at two hierarchical levels revealed low (5.2%) but significant genetic variation among populations at AFLPs (Table 3). At three hierarchical levels, a very low (0.7%) but significant proportion of the variation was among climatic zones, 4.64% was found among populations and the main proportion (94.6%) resided within populations. At two hierarchical level based on groups of populations (wild versus cultivated), variation among groups was minor (0.3%) and non-significant. The multilocus F STaflp obtained was 0.052 and significantly different from zero (Table 5). The pairwise F ST values at AFLPs (F STaflp) indicated that most of the populations were significantly differentiated although the F STaflp values were mostly low ranging from 0.005 (between Bassila and Glazoue) to 0.176 (between Zogbodomey and Segbana; data not shown). The matrices of geographic and genetic distances are correlated; the mantel test was highly significant (R = 0.383, P < 0.001; Fig. 2a). A Mantel test performed only for wild populations revealed no significant correlation (R = 0.130, P = 0.310, Fig. 2b).
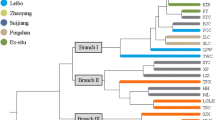
The UPGMA dendrogram based on Nei’s (1978) genetic distances revealed that some populations such as the two cultivated populations (Kand and Segb) from the north-east clustered together according to their geographic origin. Five cultivated populations clustered directly with a wild populations but not necessary with the nearest one (see, for example, NDal and Agon in Fig. 3a).
a UPGMA dendrogram representing the genetic distances among ackee populations based on Nei’s (1978) genetic distances calculated from AFLP markers; b Neighbor Joining tree of nSSR markers with the matrix of genetic distances calculated using the Cavalli-Sforza distance (Cavalli-Sforza and Edwards 1967). Numbers indicate nodes with bootstrap support in percentage for 10,000 replications. Trees constructed with populations 1.2.30 (Langella 1999). GC Guineo-Congolian, GS Guineo-Sudanian, S Sudanian, W wild population, C cultivated population
Genetic diversity and structure at nuclear microsatellites loci
The four polymorphic nuclear microsatellite markers generated a total of 22 alleles. The number of alleles per locus ranged from 2 for LMLY2 to 14 for LMLY5. The mean allelic richness (A R) ranged from 2.00 (Segbana) to 3.67 (Pobe). Observed heterozygosity values ranged from H O = 0.129 in N’Dali to H O = 0.396 in Zogbodomey. Expected heterozygosity values ranged from H E = 0.156 in N’Dali to H E = 0.419 in Savalou. Two populations had private alleles: Pobe and Glazoue with 3 and 1 alleles, respectively. The fixation index (F IS) varied among populations ranging from −0.171 in Boukombe to 0.366 in Kandi. A significant heterozygote deficit was detected only in two populations: Kandi and Pobe (Table 2).
The multilocus F STnuc (0.073) was higher than the one obtained for AFLP and significantly different from zero (Table 5). This suggested a moderate differentiation between populations that was confirmed by significant pairwise population differentiation in 24 out of 98 cases (data not shown here).
The Mantel test revealed a significant correlation between F STnuc/(1 − F STnuc) and the natural logarithm of distance (R = 0.199, P < 0.05; Fig. 2a). The Mantel test performed exclusively on wild populations was not significant (R = −0.139, P = 0.300).
The AMOVA showed that more than 92% of the total variation resided within populations (Table 4). A non-significant portion (0.65 and −0.77%) of the variation was among climatic zones and management types (wild versus cultivated), respectively. The relationships between the 14 populations based on Cavalli-Sforza distances are summarized in a neighbor-joining phylogram (Fig. 3b). Overall, the topology of the tree coincided well with the geographic distribution and the nearest populations clustered together except for three of them (Penessoulou, Bassila and Pobe). All cultivated populations are in the same subcluster together with the wild population Pobe. The bootstrap support varied between 38 and 86%.
Chloroplast microsatellite haplotypes and geographic distribution
Among the 14 primer pairs tested on a sample of 10 individuals, 12 primer pairs showed amplification products from which the product created by ccmp7 was polymorphic.
The ccmp7 primer pair was analyzed in 179 individuals; three different fragment sizes were observed: 121, 135 and 145 bp (Table 2). The two cultivated populations of the Northeast of Benin were fixed for the haplotype of 145 bp. The haplotypes of 121 and 135 bp were found in 10 and 12 populations, respectively (Fig. 1). In the six wild populations only haplotypes of 121 and 135 bp occurred and were equally frequent. The overall G ST value was 0.575 and significant (Table 5).
Multiple sequence alignments of amplification products generated by primer pairs of ccmp7 are shown in Fig. 4. High sequence homology was found (e.g., to the genus Fagus) to the atpB–rbcL intergenic region of the chloroplast genome. The microsatellite repeat motif varies for each fragment size (Table 4). The observed variation in the haplotype of 135 bp is due to a large indel of 10 bp within the mononucleotide repeat motif region, and a single-nucleotide polymorphism (SNP substitution of A by G) was observed in clone 135-1 compared to clone 135-2 (see position 116 in the sequence aligned in Fig. 4). The variation observed in haplotype 121 bp is based on 3 indels: a large indel of 12 bp within the repeat motif region of the microsatellite, an indel of one nucleotide at position 93 and an additional indel of 11 bp located in the flanking region of the microsatellite motif. For the population genetic analysis only length polymorphisms were taken into account.
Impact of farmer management practices and origin of cultivated populations
Within population genetic diversity estimates were not significantly different between the two management types (wild populations and cultivated populations) using a t-test. However, 99.2% of band richness (Br) and 96.6% of Nei’s gene diversity (H j) were kept in cultivated populations. In contrast, the cultivated populations had slightly higher values for PPL (12). Cultivated populations showed a higher percentage of non-randomly associated markers (mean = 15.0%) as compared to natural populations (11.3%; Table 2).
Comparison between the two groups of wild and cultivated populations revealed that wild populations showed in general higher values of A R and nR at nuclear microsatellites (Table 2). A R mean values for wild and cultivated populations were 3.03 and 2.57, respectively. The mean nR value for cultivated populations was 2.98 compared to 3.78 for wild populations. However, there was no significant difference between population categories according to two tailed test undertaken in FSTAT v2.9.3.2 (P = 0.131).
For the three markers, F ST or G ST estimates revealed less differentiation among wild populations compared to cultivated populations (Table 5).
Correlation between AFLP and SSR markers
Mantel’s non parametric test showed a non-significant correlation between pairwise F ST/(1 − F ST) estimates of the different markers (R = 0.165, P = 0.170; Fig. 5). Values of population pairwise F ST obtained using SSRs were not significantly higher than those derived from AFLPs (t-value = 1.218, df = 180, P = 0.225; paired t-test). This suggests that there is no meaningful correlation between AFLP and SSR diversity as assessed by the loci used in this study.
Discussion
Genetic diversity in Blighia sapida
Nei’s gene diversity within populations observed for Blighia sapida (0.14–0.18) are lower than values observed for other African indigenous fruits trees such as Adansonia digitata (0.26–0.35; Assogbadjo et al. 2006) and Uapaca kirkiana (0.22–0.32; Mwase et al. (2006) investigated with AFLP markers. Estimates derived by dominantly inherited markers (RAPD, AFLP, ISSR) are very similar and may be directly comparable (Nybom 2004). Compared with within population diversity estimates from RAPDs, B. sapida exhibits slightly lower values than Vitellaria paradoxa (0.16–0.23; Kelly et al. (2004), another important agroforestry parklands fruit species from Africa, and higher values than Melia volkensii (0.07–0.14; Runo et al. 2004). The mean population diversity is lower than RAPD mean values reported for dicotyledons (H pop = 0.191, n = 27), long-lived perennials (H pop = 0.25, n = 37), widespread (H pop = 0.22, n = 20), outcrossing (H pop = 0.27, n = 38) and late successional (H pop = 0.30, n = 16) species (Nybom and Bartish 2000; Nybom 2004).
The nSSR data indicate that B. sapida has a low level of genetic diversity compared to other forest tree species presenting similar patterns of seed dispersal and assessed with the same marker type (H E = 0.16–0.42, mean = 0.29; Tables 2 and 6). Our estimates are close to values for agroforestry fruits species undergoing domestication processes with a continental distribution such as V. paradoxa (from Mali, West Africa; Sanou et al. 2005) and Inga edulis (from Peruvian Amazon; Hollingsworth et al. 2005) (Table 6). The mean within-population diversity (H E = 0.286) is also lower than SSR mean values reported for long-lived perennial (H E = 0.68, n = 59), widespread (H E = 0.62, n = 31), outcrossing (H E = 0.65, n = 71) and late successional species (H E = 0.70, n = 34; Nybom 2004). The multilocus fixation index F IS for the total population as well as for each group of wild and cultivated populations was not significantly different from zero. Only populations Kandi and Pobe deviated from HWE. During the primers characterization (Ekué et al. 2009b), locus LMLY5 in Kandi showed evidence for the presence of a null allele (excess of homozygotes for most allele size classes) which could explain the heterozygote deficit for this population. Significantly positive F IS value has been attributed to population substructure and inbreeding (Kelly et al. 2004; Ueno et al. 2005) which might be the case in the Pobe population. Although the effect of the sampling variance could be high because our sample sizes were low (<30) in many populations and could have impacted the F IS estimation, B. sapida appears to be predominantly outcrossing.
Only one chloroplast microsatellite marker (ccmp7) was polymorphic with three different haplotypes. The large difference in size variation observed between the three haplotypes results probably from long time mutation among them. More sampling across the whole West Africa could lead to the discovery of new haplotypes.
Human activity and genetic diversity within cultivated populations of Blighia sapida
The domestication process in Blighia sapida resulted in a slight non-significant reduction in genetic variation in the cultivated populations as compared to the wild populations at both AFLP and nSSR markers. Artificial selection during domestication often results in a progressive narrowing of the genetic base of the cultivated populations. However, perennial domesticated species usually retain relatively higher percentages of the variation of their wild ancestors compared to annual plants (Tanksley and McCouch 1997; Miller and Schaal 2006; Pickersgill 2007). For example, nuclear microsatellite studies revealed that cultivated columnar cacti (Polaskia chichipe) populations retained 88–96% of the variation of their ancestors (Otero-Arnaiz et al. 2005). The shea tree (V. paradoxa) subjected to different human management intensities in field/fallow stands kept 95% of the genetic diversity present in forest stands (Kelly et al. 2004); while cultivated populations of the tropical fruit tree I. edulis and their wild ancestors did not differ in gene diversity (Hollingsworth et al. 2005). AFLPs markers showed that populations of the cultivated tropical fruit tree Spondias purpurea retained approximately 90% of the variation detected in wild populations. The reduction in genetic diversity (nearly 5% for AFLPs and 15% for nSSRs) observed for B. sapida are somehow similar or even lower, which is consistent with the reduction in variation seen in other long-lived species. In addition, there was a high proportion (1.4 times more) of non-randomly associated loci in cultivated populations than in wild populations observed at AFLP markers. Thus, farmer-led domestication activities slightly reduced the genetic diversity of ackee in Benin. However, this did not strongly affect diversity within populations, since effective population sizes were kept high enough to avoid strong losses of genetic diversity due to drift.
Domestication affects the structure of genetic variation in cultivated populations
The AMOVA results for AFLPs agree with general observations that perennial and outbreeding species with wide and more or less continuous distribution maintain most of their variation within populations (Hamrick et al. 1992). According to Wright’s (1978) guidelines for interpretation of F STaflp values, 0.052 suggests moderate differentiation and is consistent with the life history of B. sapida (Ekué et al. 2004). Similar low levels of AFLP population differentiation were observed for another outcrossing African indigenous fruits tree species Uapaca kirkiana (6.8%; Mwase et al. 2006). This result is in accordance with the general tendency of tropical trees to possess most of their genetic diversity within populations (Hamrick and Loveless 1989).
With nSSRs, even though most of the genetic diversity is contained within populations (F STnuc: 7% for all populations and 5% for wild populations), the populations can be considered to be genetically distinct. In accordance with its continental distribution, B. sapida is expected to have a lower F STnuc value compared to insular species (Muller et al. 2009) as shown in Table 6. The genetic differentiation among populations of B. sapida is comparable to species with a continental distribution and possessing the same seed dispersal mode (e.g., V. paradoxa and I. edulis, Table 6).
Domestication impacts not only the amount of genetic variation contained in cultivated populations, but also the structure of this variation. Hamrick and Godt (1997) reported that the mean G ST level (analogous to Φ ST and F ST) for crop species as 0.339, while the mean G ST for noncrop species is 0.212. Our results are consistent with this trend: cultivated B. sapida populations have a greater portion of their variance (measured as Φ ST, F ST or G ST; see Tables 3 and 5) distributed among populations than wild B. sapida populations. In addition, cultivated populations showed 1.5 times higher levels of inbreeding compared to wild populations. We infer that the higher differentiation among cultivated B. sapida populations relative to their wild ancestors reflects human influences on the reproductive system in cultivated populations. Indeed, quantitative surveys on indigenous knowledge and traditional management of B. sapida in Benin revealed that farmers practiced assisted tree regeneration, transplantation and sowing of seeds of trees having their preferred fruit traits. In addition, traditional silvicultural practices such as ringing, grazing protection, pruning, fire protection are used only for ‘cultivated’ trees (Ekué et al. 2009b, 2010). This selection against undesirable phenotypes is likely changing the genetic structure in cultivated populations. Since there is variation of the indigenous knowledge and management practices among ethnic groups (Ekué et al. 2009b, 2010), the changes of the genetic structure within each cultivated population are different and likely leading to the differentiation among them. The differentiation of the two cultivated populations in the North-east completely fixed for haplotype of 145 bp absent from the wild populations is an indication that after initial establishment of each cultivated population, human activities appear to be regionally limited and have not promoted extensive gene flow over long-distance.
The spatial clustering of wild populations is likely facilitating gene flow among them resulting in a low differentiation. This argument is supported by the absence of isolation by distance patterns obtained with the Mantel test performed only on wild populations at both AFLPs and nSSRs markers.
Non-congruence among AFLPs and SSRs marker classes of population differentiation
Poor correlation between estimates of genetic similarity based on AFLP and SSR suggests that both markers may selectively screen complementary, rather than overlapping, regions of the genome (Crouch et al. 1999). Thus, it is useful to utilize a range of marker systems in order to generate highly accurate estimates of genetic similarity in genetic diversity studies of B. sapida.
Origin of Blighia sapida cultivated populations
Although wild populations and cultivated populations do not match geographically (making it more difficult to determine precisely the origin of cultivated populations), ccmp7 and AFLP data provided evidence that most cultivated populations of B. sapida have probably originated mainly from wild populations in Benin. However, the haplotype of 145 bp present in the two cultivated populations restricted to the northeast is completely absent from wild populations. This is an indication that gene flow by seeds is occurring from neighboring countries, most likely from Nigeria in this case. However, the clustering of each of the remaining five cultivated populations with different wild populations suggests their geographic origin in Benin. Thus, our data strongly indicate that cultivated B. sapida is not a homogenous entity in Benin.
Implications for conservation and sustainable use
The domestication process in B. sapida in Benin has slightly reduced levels of genetic variation in cultivated populations as compared to their wild progenitors. Cultivated populations from different climatic zones have different origins and contain varying levels of genetic variation. The proportion of genetic variation distributed among populations is greater in cultivated populations than in wild populations. Germplasm sources for establishment of new planted trees appear to be limited to trees already growing in cultivated populations. Both wild and cultivated populations of ackee are therefore valuable gene pools for conservation and future breeding.
A multiple breeding population strategy (Namkoong 1989) is recommendable and will promote the sustainable use while ensuring the circa situ conservation of the genetic diversity currently present within cultivated populations. Farmers must be trained to maintain good collection practices of currently cultivated ackee germplasm as they regenerate the species in subsequent cycles of mating and selection of on-farm propagation (Dawson et al. 2008). To extend domestication activities, exchange of germplasm should be limited to neighboring farms and villages. As some populations are highly differentiated, any transfer of germplasm between such populations may not be the best mean to promote on farm conservation, because of possible productivity losses through outbreeding depression and genetic dilution (Jamnadass et al. 2005).
For in situ conservation, representative units of conservation should be defined in each climatic zone. B. sapida may be considered as common or not (depending on different localities), as exploited in different intensity and for different purposes across the country, and its habitat as secure or not. In the Guinean zone in Southern Benin, the species is less used and all populations found there are natural. Since 60% of Benin’s population occurs in this area, there is a big pressure on land resources for agricultural production. Therefore, representative units for conservation in this zone should target populations located next to already existing protected areas. In the Guineo-Sudanian zone, the two natural populations surveyed are located in protected areas and can be considered as conservation units. In the Sudanian zones, conservation units should target populations in the Western block (near Togo) and the Eastern block (near Nigeria) in order to maximize genetic diversity as the origin of the later block is likely to be different from other populations in Benin.
For an eventual establishment of provenance trials, seeds must be collected also from all representative units including cultivated populations. Since the heritability of important characters including fruit traits is unknown, we recommend also establishing progeny trials with seeds collected from selected mother trees.
Blighia sapida is native from West Africa and results from this study suggested the presence of haplotypes that are predominant in neighboring countries like Nigeria. Therefore, further studies are suggested to explore genetic diversity in wild and managed populations at molecular markers, adaptive traits, and traits of economic importance based on a wider sampling across the whole distribution range including transferred land races and modeling on longer cycles.
References
Assogbadjo AE, Kyndt T, Sinsin B, Gheysen G, Van Damme P (2006) Patterns of genetic and morphometric diversity in baobab (Adansonia digitata) populations across different climatic zones of Benin (West Africa). Ann Bot 97:819–830
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Am J Hum Genet 19:233–257
Clement C (1999) 1492 and the loss of amazonian crop genetic resources. ii. crop Biogeography at contact. Econ Bot 53:203–216
Coart E, Glabeke SV, Petit RJ, Bockstaele EV, Roldán-Ruiz I (2005) Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.). Conserv Genet 6:259–273
Codjia JTC, Assogbadjo AE, Ekué MRM (2003) Diversité et valorisation au niveau local des ressources végétales forestières alimentaires du Bénin. Cahiers Agricultures 12:321–331
Collevatti RG, Grattapaglia D, Hay JD (2001) Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Mol Ecol 10:349–356
Crouch JH, Crouch HK, Constandt H, Van Gysel A, Breyne P, Van Montagu M, Jarret RL, Ortiz R (1999) Comparison of PCR-based molecular marker analyses of Musa breeding populations. Mol Breeding 5:233–244
Dawson I, Hollingsworth P, Doyle J, Kresovich S, Weber J, Sotelo Montes C, Pennington T, Pennington R (2008) Origins and genetic conservation of tropical trees in agroforestry systems: a case study from the Peruvian Amazon. Conserv Genet 9:361–372
Deguilloux MF, Dumolin-Lapègue S, Gielly L, Grivet D, Petit RJ (2003) A set of primers for the amplification of chloroplast microsatellites in Quercus. Mol Ecol Notes 3:24–27
Dossou MKR, Codjia JTC, Biaou G (2004) Rôle de la ressource forestière Blighia sapida (ackee ou faux acajou) dans l’économie des ménages du Nord-Ouest du Bénin. Bulletin de la Recherche Agronomique du Bénin 46:33–41
Dutech C, Joly HI, Jarne P (2004) Gene flow, historical population dynamics and genetic diversity within French Guianan populations of a rainforest tree species, Vouacapoua americana. Heredity 92:69–77
Ekué MRM, Assogbadjo AE, Mensah GA, Codjia JTC (2004) Aperçu sur la distribution écologique et le système agroforestier traditionnel autour de l’ackée (Blighia sapida) en milieu soudanien au Nord Bénin. Bulletin de la Recherche Agronomique du Bénin 44:34–44
Ekué MRM, Gailing O, Finkeldey R (2009a) Transferability of Simple Sequence Repeat (SSR) Markers Developed in Litchi chinensis to Blighia sapida (Sapindaceae). Plant Molecular Biology Reporter 27:570–574
Ekué MRM, Gailing O, Finkeldey R, Eyog-Matig O (2009b) Indigenous knowledge, traditional management and genetic diversity of the endogenous agroforestry species ackee (Blighia sapida) in Benin. Acta Hortic 806:655–661
Ekué MRM, Sinsin B, Eyog-Matig O, Finkeldey R (2010) Uses, traditional management, perception of variation and preferences in ackee (Blighia sapida K.D. Koenig) fruit traits in Benin: implications for domestication and conservation. J Ethnobiol Ethnomed 6:12
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Emshwiller E, Theim T, Grau A, Nina V, Terrazas F (2009) Origins of domestication and polyploidy in oca (Oxalis tuberosa; Oxalidaceae). 3. AFLP data of oca and four wild, tuber-bearing taxa. American Journal of Botany 96:1839–1848
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes - Application to Human Mitochondrial-DNA Restriction Data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2007) ARLEQUIN ver 3.11. An integrated software package for population genetics data analysis. Computational and Molecular Population Genetics Lab (CMPG), University of Berne, Switzerland. http://cmpg.unibe.ch/software/arlequin3. Accessed 15 November 2009
Felsenstein J (1993) PHYLIP: phylogenetic inference package. Version. 3.5. Department of Genetics, University of Washington, Seattle, USA
Fontaine C, Lovett PN, Sanou H, Maley J, Bouvet JM (2004) Genetic diversity of the shea tree (Vitellaria paradoxa C.F. Gaertn), detected by RAPD and chloroplast microsatellite markers. Heredity 93:639–648
Gepts P (2004) Crop domestication as a long-term selection experiment. Plant Breeding Reviews 24:1–44
Goudet J (2001) FSTAT: A program to estimate and test gene diversities and fixation indices, (Version 2.9.3.2). University of Lausanne, Switzerland
Goudet J, Raymond M, deMeeus T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis. http://www.mbio.ncsu.edu/BioEdit/bioedit.html. Accessed 15 November 2009
Hamrick JL, Loveless MD (1989) The genetic structure of tropical tree populations: associations with reproductive biology. In: Bock JH, Linhart YB (eds) The evolutionary ecology of plants. Westview Press, Boulder, pp 129–146
Hamrick JL, Godt MJW (1997) Allozyme diversity in cultivated crops. Crop Sci 37:26–30
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests 6:95–124
Hollingsworth PM, Dawson IK, Goodall-Copestake WP, Richardson JE, Weber JC, Sotelo Montes C, Pennington RT (2005) Do farmers reduce genetic diversity when they domesticate tropical trees? A case study from Amazonia. Mol Ecol 14:497–501
ICRAF (International Centre for Research in Agroforestry) (2009) Available from http://www.worldagroforestry.org/Sites/TreeDBS/aft/speciesPrinterFriendly.asp?Id=17953. (Accessed 15 November 2009)
Jamnadass R, Hanson J, Poole J, Hanotte O, Simons TJ, Dawson IK (2005) High differentiation among populations of the woody legume Sesbania sesban in sub-Saharan Africa: Implications for conservation and cultivation during germplasm introduction into agroforestry systems. For Ecol Manag 210:225–238
Kelly BA, Hardy O, Bouvet JM (2004) Temporal and spatial genetic structure of Vitellaria paradoxa (shea tree) in an agroforestry system in southern Mali. Mol Ecol 13:1231–1240
Langella O (1999) POPULATIONS 1.2.30 Population genetic software. http://bioinformatics.org/~tryphon/populations. Accessed 15 November 2009
Lemes MR, Gribel R, Proctor J, Grattapaglia D (2003) Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon, based on variation at microsatellite loci: implications for conservation. Mol Ecol 12:2875–2883
Lewis PO, Zaykin D (2001) Genetic Data Analysis: Computer program for the analysis of allelic data. Version 1.0 (d16c). Free program distributed by the authors over the internet from http://lewis.eeb.uconn.edu/lewishome/software.html. Accessed 15 November 2009
Lhuillier E, Butaud J-F, Bouvet J-M (2006) Extensive clonality and strong differentiation in the insular pacific tree Santalum insulare: implications for its conservation. Ann Bot 98:1061–1072
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mantel N (1967) The Detection of Disease Clustering and a Generalized Regression Approach. Cancer Res 27:209–220
Miller AJ, Schaal BA (2006) Domestication and the distribution of genetic variation in wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea L. (Anacardiaceae). Mol Ecol 15:1467–1480
Muller F, Voccia M, Bâ A, Bouvet J (2009) Genetic diversity and gene flow in a Caribbean tree Pterocarpus officinalis Jacq.: a study based on chloroplast and nuclear microsatellites. Genetica 135:185–198
Mwase WF, Bjørnstad Å, Stedje B, Bokosi JM, Kwapata MB (2006) Genetic diversity of Uapaca kirkiana Muel. Årg. Populations as revealed by amplified fragment length polymorphisms (AFLPs). Afr J Biotechnol 5:1205–1213
Namkoong G (1989) Population genetics and the dynamics of conservation. In: Knutson L, Stoner AK (Eds). Biotic diversity and germplasm preservation, global imperatives. Kluwer Academic Publishers. ISBN 0-7923-0178-1. pp 161-181
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3:93–114
Otero-Arnaiz A, Casas A, Hamrick JL, Cruse-Sanders J (2005) Genetic variation and evolution of Polaskia chichipe (Cactaceae) under domestication in the Tehuacán Valley, central Mexico. Mol Ecol 14:1603–1611
Peakall R, Smouse PE (2006) GenAlEx 6.2: Genetic Analysis in Excel. Population genetic software for teaching and research. The Australian National University, Canberra, Australia. http://www.anu.edu.au/BoZo/GenAlEx/
Pen M (Jamaica Information Service) (2006) Viable Ackee Industry must be Protected- BSJ Inspector. www.jis.gov.jm/agriculture/html/20060506T100000-0500_8777_JIS_VIABLE_ACKEE_INDUSTRY_MUST_BE_PROTECTED__BSJ_INSPECTOR.asp. Accessed 15 April 2009
Petit RJ, El Mousadik A, Pons O (1998) Identifying Populations for Conservation on the Basis of Genetic Markers. Conserv Biol 12:844–855
Pickersgill B (2007) Domestication of Plants in the Americas: Insights from Mendelian and Molecular Genetics. Ann Bot 100:925–940
Rice WR (1989) Analysing tables of statistical test. Evolution 43:223–225
Rousset F (1996) Equilibrium Values of Measures of Population Subdivision for Stepwise Mutation Processes. Genetics 142:1357–1362
Runo MS, Muluvi GM, Odee DM (2004) Analysis of genetic structure in Melia volkensii (Gurke) populations using random amplified polymorphic DNA. Afr J Biotechnol 38:421–425
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Sanou H, Lovett PN, Bouvet JM (2005) Comparison of quantitative and molecular variation in agroforestry populations of the shea tree (Vitellaria paradoxa C.F. Gaertn) in Mali. Mol Ecol 14:2601–2610
Sim CH, Mahani MC, Choong CY, Salma I (2005) Transferability of SSR markers from lychee (Litchi chinensis Sonn.) to pulasan (Nephelium ramboutan-ake L.). Fruits 60:379–385
Tanksley SD, McCouch SR (1997) Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild. Science 277:1063–1066
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ueno S, Setsuko S, Kawahara T, Yoshimaru H (2005) Genetic diversity and differentiation of the endangered Japanese endemic tree Magnolia stellata using nuclear and chloroplast microsatellite markers. Conserv Genet 6:563–574
Vekemans X (2002) AFLP-SURV version 1.0 Distributed by the author. Laboratoire de génétique et Ecologie Végétale, Univ. Libre de Bruxelles, Belgium
Viruel MA, Hormaza JI (2004) Development, characterization and variability analysis of microsatellites in lychee (Litchi chinensis Sonn., Sapindaceae). Theor Appl Genet 108:896–902
Vos P, Hogers R, Bleeker M, Reijans M, Tvd Lee, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the Analysis of Population-Structure. Evolution 38:1358–1370
Weising K, Gardner RC (1999) A set of conserved PVR primers for the analysis of simple sequence repeat polymorphism in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Woodhead M, Russell J, Squirrell J, Hollingsworth PM, Mackenzie K, Gibby M, Powell W (2005) Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Mol Ecol 14:1681–1695
Wright S (1978) Evolution and genetics of populations: Variability within and among natural populations, vol 4. University of Chicago Press, Chicago, USA
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgments
This research was supported by grant and fellowships from the International Foundation for Science, Rothamsted International and Bioversity International to the first author. We are grateful to Hugues Akpona for field assistance and we appreciate the laboratory assistance of Oleksandra Dolynska.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ekué, M.R.M., Gailing, O., Vornam, B. et al. Assessment of the domestication state of ackee (Blighia sapida K.D. Koenig) in Benin based on AFLP and microsatellite markers. Conserv Genet 12, 475–489 (2011). https://doi.org/10.1007/s10592-010-0155-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0155-z