Abstract
Bumblebees (Bombus spp.) are commonly used for greenhouse pollination of tomatoes and other crop plants. The colonies used for this purpose are provided by commercial bumblebee breeders, which by now operate at a professional company level. As a result of this practice commercially bred bumblebee colonies are transported and used over large distances and national borders, introducing subspecies into non-endemic regions. The question whether and to what extends gene flow between such managed greenhouse and wild bumblebee populations exists, so far has not been addressed. Here we used samples from three greenhouses in Poland and the surrounding populations to address this question. Using microsatellite DNA data we found strong genetic introgression from the sampled greenhouse populations into the adjacent populations. Depending on the analysed population, the number of individuals assigned to the greenhouse populations ranged from 0.08 to 0.47. We also found that more distant populations were much less affected by genetic introgression from the greenhouses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bumblebees (Bombus spp.) are increasingly used in greenhouses for pollination services, because they are very efficient pollinators that can be handled with great ease. Bumblebees exceed other pollinator species in pollination efficiency, due to their behavioural particularity of “buzz pollination” (Buchmann 1985) especially for many Solanaceae, including tomatoes (Solanum lycopersicum) or bell peppers (Capsicum annuum). An additional advantage for their commercial use in greenhouses is that colonies can be easily bred, handled and transported throughout the year. Colonies can be delivered within a few days on demand, which makes them a perfect insect for indoor pollination. Bumblebee colonies became commercially available for greenhouse pollination from 1980s, and the species of choice in Europe was the buff-tailed bumblebee B. terrestris, one of the most common and widespread European species. Even though B. terrestris is not the only species used in greenhouse pollination (e.g. B. impatiens is the used species in North and Central America) it is definitely the dominant one on the market and is exported to Asia, Australia and South America. The total number of bumblebee colonies reared for greenhouses in 2006 was estimated to be nearly as high as one million (Velthuis and van Doorn 2006).
The downside of the massive exports of bumblebee colonies from Europe into non-endemic areas is that B. terrestris has established stable feral populations in New Zealand, Japan, Australia, Tasmania, Chile and Israel (Buttermore 1997; Goulson 2003; Goulson and Hanley 2004; Nagamitsu and Yamagishi 2009; Williams et al. 2009; Williams and Osborne 2009). This quite impressively shows the colonizing capacity of B. terrestris which may be facilitated by male haploidy which exposes all recessive alleles to selection (Schmid-Hempel et al. 2007). Whatever the reason for this evolutionary success story of B. terrestris, the introduction of this bumblebee species to non-endemic regions has raised severe concerns from a conservation perspective. Inoue et al. (2008) report that B. terrestris may displace native bumblebee species and the worldwide shipping of colonies may also cause the worldwide spread of their diseases (Goka et al. 2001, 2006) spilling over from greenhouses into wild populations (Colla et al. 2006; Otterstatter and Thomson 2008).
Given these negative effects of B. terrestris exportations to non-endemic regions, what is to be expected for Europe, where it is an endemic species? Clearly, from a conservation point of view it would be optimal if bumblebee stocks used in greenhouse pollination were descendants of local populations (Velthuis and van Doorn 2006) to avoid potential negative effects on endemic pollinator communities. Unfortunately this is generally not the case in commercial bumblebee rearing. In fact, the European B. terrestris market is dominated by the use of two southern subspecies namely B. t. dalmatinus or B. t. sassaricus, which have proved to be the most suitable for colony rearing. In spite of evidence that bumblebee workers can fly out of greenhouses (Whittington et al. 2004) many European authorities still assume that it is unlikely that males or young queens escape from greenhouses. And even if bumblebees might escape, they also assume that the lack of synchronization with local populations would prevent any interbreeding. Only a few countries (Norway, Turkey, Israel and Spain for Canary Islands) in Europe have imposed import restrictions on non-endemic bumblebee species (Velthuis and van Doorn 2006). In the light of experiences with the invasion of imported bumblebees in Japan (Inoue et al. 2008), there seems to be reason for a more cautious policy of bumblebee usage in greenhouse pollination. Today it is unclear to what extend greenhouse populations interfere with and genetically alters the surrounding wild bumblebee populations. Here we use a population genetic approach to study three selected greenhouse populations of B. terrestris and their surrounding wild populations in Southern Poland, to test for a potential introgression of commercial greenhouse bumblebee populations into the wild.
Materials and methods
Sampling
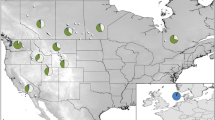
We sampled a total of 588 B. terrestris workers from nine populations in Southern Poland (Map 1). Three of these populations were directly collected in greenhouses and thus being of imported, non-endemic origin. Three endemic populations were sampled in the area surrounding the greenhouses and three more endemic populations without any close contact to greenhouses at a distance of at least 30 km away. The sample sizes, names, GPS data and abbreviations of the populations are given in Table 1. All sampled workers were stored in 95% ethanol at −20°C until DNA extraction.
Genotyping
Total DNA was extracted from one leg of each of the sampled workers following a modified Chelex extraction protocol as described by Walsh et al. (1991). All individuals were genotyped at four microsatellite loci (B11, B96, B124, B126; Estoup et al. 1993, 1995) following standard polymerase chain reaction (PCR) protocols in automated DNA capillary sequencers (MegaBACE 1000) according to manufacturer’s instructions.
Population genetic analysis
Before embarking on classical population genetic analyses, it is important to assign each individual worker to its putative mother colony resulting in a mother and father genotype for each colony. This way biases due to sampling multiple workers of the same colony can be avoided. Queen and father drone genotypes were inferred using the COLONY 1.3 algorithm (Wang 2004) performing four runs with different seed numbers and using those genotype assignments with the highest log probability for further analyses. Based on the mother/father genotypes we calculated the observed and expected heterozygosities, deviation from Hardy–Weinberg equilibrium, pairwise F ST values and population differentiation, using the Excel Microsatellite Toolkit 3.1.1 (Park 2001) and Genepop (Version: Genepop on the Web; Raymond and Rousset 1995). The number of alleles and the allelic richness was calculated using HP-Rare 2.0 (Kalinowski 2004, 2005).
To infer the degree of introgression, we determined the number of individuals sampled in the surrounding populations, but which were assigned to the greenhouse populations. The individual workers were assigned using BAPS 2.0 (Corander et al. 2004). We used the trained clustering option of the software (Corander et al. 2008) with the greenhouse populations as fixed clusters of known origin. Since two of the populations (K-GH and P-GH) were originating from the same supplier they were pooled in one cluster, while the remaining greenhouse population S-GH formed the second fixed cluster. All other populations were treated as of unknown origin, thus it was possible to test which of the endemic populations' colonies were clustered to one of the greenhouse clusters. To infer the degree of introgression we used ten replicate runs on BAPS, using the mean and standard deviation of the number of individuals assigned to the greenhouses as our final estimate of introgression.
Results
Based on the sample of 588 workers from the nine populations we inferred a total of 161 colonies, each represented by one queen and male genotype. The number of colonies inferred for each population, the number of alleles and the allelic richness in the parental population (based on male and queen genotypes) are given in Table 1. The observed heterozygosities ranged from 0.57 in the K-GH population up to 0.87 in the S-C population, all observed and expected heterozygosity values are given in Table 1. Only one population (K) deviated significantly from Hardy–Weinberg proportions, which however did not remain significant after Bonferroni correction for multiple tests. The allelic richness over all loci ranged from 6.7 ± 1.6 (mean ± SD) in the K-GH population up to 8.8 ± 3.5 in the P population, all values of the observed number of alleles and the allelic richness are given in Table 1. The pairwise F ST values ranged from 0.02 to 0.22, with the lowest value for the two greenhouse populations provided by the same commercial bumblebee breeder. The test for population differentiation revealed that only 4 out of the 36 pairwise tests showed no significant population differentiation after Bonferroni correction for multiple tests. These were the Populations Kr-Gh/P-Gh, S-Gh/S, K/P and P/S. The F ST values and the significance of the Fisher’s exact test for population differentiation are given in Table 2.
The estimated degree of individuals assigned to the greenhouse populations ranged from 0.08 in the K-C and S-C populations upto 0.47 in the S population. Moreover the more distant control populations showed significantly lower average amount of individuals assigned to the greenhouses (0.13 ± 0.08) than the close by populations (0.38 ± 0.08). The detailed percentages of individuals assigned to the greenhouse populations are given in Table 1.
Discussion
In this study we estimated the degree of genetic introgression from Bombus terrestris greenhouse populations into feral populations by assigning individuals to their potential population of origin. The percentage of individuals assigned to the greenhouse populations was very high in the nearby wild populations surrounding the greenhouses. The percentage of introgression ranged from 33 to 47% and thus was significantly higher than the percentage estimated for the more distant control populations, which however still had between 8 and 21% of individuals assigned to the greenhouse populations. When taking a closer look at the introgression pattern in the nearby wild populations, the degree of introgression corresponds to the greenhouse population they are neighbouring. The populations K and P have a much higher degree of introgression from the K-GH/P-GH population complex than from the S-GH population, while the opposite is true for the S population which neighbours to the S-GH population (Table 1).
The test for population differentiation revealed the K-GH and P-GH populations did not differ significantly from each other, which is expected, since the bumblebees used in these two greenhouses are from the same supplier, and are thus from a similar or the same breeding stock. Also within the group of the nearby populations, K, P and S, three pairwise comparisons were not significant, even though there is no obvious consistent pattern of genetic differentiation. The high similarity among the populations may result from repeated introgressions from the greenhouse populations in the past to such a degree that three of the pairwise tests for genetic differentiation fail to detect any sub-structuring. Another reason for the three non significant pairwise tests might also be a lack of power due to our usage of four microsatellite markers. A higher number of markers might have resulted in a better resolution and would thus be desirable for future studies.
The pairwise FST values provide a similar picture where the three greenhouse populations show the lowest degree of genetic differentiation (average FST = 0.05), with the lowest of all detected values (FST = 0.02) between the two populations from the same supplier, the populations K-GH and P-GH. Substructuring is however also low among the K, P and S populations with an average FST = 0.06. The highest pairwise FST values were found between the control populations, A-C, S-C and K-C (average FST = 0.15), with K-C as the genetically most differentiated population to all other ones (average FST = 0.19).
Interestingly the two populations with lowest allelic richness and the lowest value for the observed heterozygosity are the two populations K-GH and P-GH populations. While this might be a sign of inbreeding due to the breeding of bumblebees from a limited number of queens, there is no deviation from Hardy–Weinberg and our study was generally not designed to detect such effects. So, it remains difficult to judge whether the commercial breeding had already an inbreeding effect on the greenhouse populations.
We used an assignment test based on trained clustering to infer the percentage of individuals in the populations for the estimation of genetic introgression. The genetic characteristics suggest that introgressed individuals are direct descendents from the greenhouse populations, either offspring of gynes which had escaped from the greenhouses or workers which were conducting foraging flights outside the greenhouses. Although we can certainly not exclude the latter we expect this to be a rare phenomenon since there is ample forage in the greenhouses. Worker bees typically forage at nearby food sources and not necessarily leave the greenhouse. In the more distant control populations this phenomenon can be excluded because they are outside the foraging range of the greenhouse colonies which does normally not exceed 0.8 km (Wolf and Moritz 2008).
The degree of introgression in the more distant populations was as low as 7% and thus markedly lower than around the greenhouses. However these populations were chosen under the assumption to be not affected by introgression. Therefore we cannot exclude that the estimated introgression is an artefact of the estimation procedure, which would mean that also our estimates for the nearby populations are overrated. However even in this case where we would assume an average false introgression rate of 12.1% (which is the average for the distant populations), our estimates for the nearby populations would still range between 20 and 35%. In contrast, if we consider the genetic introgression results for the distant populations as valid, they imply that genes from the greenhouse populations already have spread much further than initially expected. This definitely would be the worst case scenario from a conservation perspective.
Regardless of scenario or statistical pitfalls, our results strongly suggest that greenhouse bumblebees spread their genes into feral conspecific populations. This is in line with previous studies, which already showed that imported greenhouse bumblebees can become an invasive species in competition with native species (e.g. Goulson and Hanley 2004; Hingston 2006; Inoue et al. 2008; Williams and Osborne 2009). In our case of conspecific introgression, the effects and consequences of genetic spillover from non-endemic subspecies to endemic ones are harder to predict. The genetic introgression will surely alter the genetic structure of a given population, which in turn may lead to the loss of specific traits and local adaptations. Introgression will depend on the scale of import and the ratio of wild to greenhouse colonies. Today we do not know the tolerable threshold value of influx of non-endemic genes to maintain stable local population.
In conclusion our results imply that bumblebees used in greenhouses can not only trigger a classical biological invasion but are also capable of changing the genetic structure of conspecific endemic subspecies. Since the consequences of this genetic spillover are hard to predict and might have potential negative affects for the conservation efforts of bumblebees, it seems prudent to handle and manage bumblebee imports with caution in regions where B. terrestris is endemic.
References
Buchmann SL (1985) Bees use vibration to aid pollen collection from non-poricidal flowers. J Kansas Entomol Soc 58:517–525
Buttermore RE (1997) Observations of successful Bombus terrestris (L.) (Hymenoptera: Apidae) colonies in southern Tasmania. Aust J Entomol 36:251–254
Colla SR, Otterstatter MC, Gegear RJ, Thomson JD (2006) Plight of the bumble bee: pathogen spillover from commercial to wild populations. Biol Conserv 129:461–467
Corander J, Waldmann P, Marttinen P, Sillanpää MJ (2004) BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20:2363–2369
Corander J, Marttinen P, Sirén J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539. doi:10.1186/1471-2105-9-539
Estoup A, Solignac M, Harry M, Cornuet JM (1993) Characterization of (Gt)N and (Ct)N microsatellites in 2 insect species – Apis mellifera and Bombus terrestris. Nucleic Acid Res 21:1427–1431
Estoup A, Scholl A, Pouvreau A, Solignac M (1995) Monoandry and polyandry in bumble bees (Hymenoptera - Bombinae) as evidenced by highly variable microsatellites. Mol Ecol 4:89–93
Goka K, Okabe K, Yoneda M (2001) Bumblebee commercialization will cause worldwide migration of parasitic mites. Mol Ecol 10:2095–2099
Goka K, Okabe K, Yoneda M (2006) Worldwide migration of parasitic mites as a result of bumblebee commercialization. Pop Ecol 48:285–291. doi:10.1007/s10144-006-0010-8
Goulson D (2003) Effects of introduced bees on native ecosystems. Annu Rev Ecol Evol S 34:1–26
Goulson D, Hanley ME (2004) Distribution and forage use of exotic bumblebees in South Island, New Zealand. New Zeal J Ecol 28:225–232
Hingston AB (2006) Is the exotic bumblebee Bombus terrestris really invading Tasmanian native vegetation? J Insect Conserv 10:289–293
Inoue MN, Yokoyama J, Washitani I (2008) Displacement of Japanese native bumblebees by the recently introduced Bombus terrestris (L.) (Hymenoptera : Apidae). J Insect Conserv 12:135–146
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kalinowski ST (2005) HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Nagamitsu T, Yamagishi H (2009) Nest density, genetic structure, and triploid workers in exotic Bombus terrestris populations colonized Japan. Apidologie 40:429–440
Otterstatter MC, Thomson JD (2008) Does pathogen spillover from commercially reared bumble bees threaten wild pollinators? PLoS ONE 3:e2771. doi:10.1371/journal.pone.0002771
Park SDE (2001) Trypanotolerance in West African cattle and the population genetic effects of selection (PhD. thesis), University of Dublin
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Schmid-Hempel P, Schmid-Hempel R, Brunner PC, Seeman OD, Allen GR (2007) Invasion success of the bumblebee, Bombus terrestris, despite a drastic genetic bottleneck. Heredity 99:414–422
Velthuis HHW, van Doorn A (2006) A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 37:421–451
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–512
Wang J (2004) Sibship reconstruction from genetic data with typing errors. Genetics 166:1963–1979
Whittington R, Winston ML, Tucker C, Parachnowitsch AL (2004) Plant-species identity of pollen collected by bumblebees placed in greenhouses for tomato pollination. Canad J Plant Sc 84:599–602
Williams PH, Osborne JL (2009) Bumblebee vulnerability and conservation world-wide. Apidologie 40:367–387
Williams P, Colla S, Xie ZH (2009) Bumblebee vulnerability: common correlates of winners and losers across three continents. Conserv Biol 23:931–940
Wolf S, Moritz RFA (2008) Foraging distance in Bombus terrestris L. (Hymenoptera: Apidae. Apidologie 39:419–427. doi:10.1051/apido:2008020
Acknowledgments
We would like to thank P. Leibe for assistance and help with the laboratory work. This study was funded by the EU-Project ALARM (Assessing Large scale environmental Risks for biodiversity with tested Methods), Pollinator Module, EU Contract No.: 506675).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kraus, F.B., Szentgyörgyi, H., Rożej, E. et al. Greenhouse bumblebees (Bombus terrestris) spread their genes into the wild. Conserv Genet 12, 187–192 (2011). https://doi.org/10.1007/s10592-010-0131-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0131-7