Abstract
An emerging pattern is that population densities of generalist rodents are higher in small compared to large forest patches in fragmented landscapes. We used genetically based measures of migration between patches to test two dispersal-based hypotheses for this negative density-area relationship: (1) emigration rates from small patches should be relatively lower compared to large patches (“inhibited dispersal hypothesis”), or (2) immigration rates should be higher into small than large patches (“immigration hypothesis”). Neither hypothesis was supported using data on dispersal inferred from eight microsatellite loci for 12 populations of Peromyscus leucopus in six small (1.3–2.7 ha) and six large (8–150 ha) forest patches. Emigration rates were not lower from and immigration rates were not higher into small than large patches. In fact, contrary to both hypotheses, emigration rates were higher from populations of P. leucopus in small compared to large patches. Based on a combination of genetic and field data, we speculate that higher reproduction in smaller patches resulted in higher densities which led to higher emigration rates from those patches. Rates of reproduction (presumably driven by better habitat conditions in smaller patches), rather than dispersal, seems to drive density differences in forest patches. We conclude that smaller forest patches within an agricultural matrix act as a source of individuals, and that migration rates are fairly high among forest patches regardless of size.



Similar content being viewed by others
References
Abramsky Z, Van Dyne GM (1980) Field studies and a simulation model of small mammals inhabiting a patchy environment. Oikos 35:80–92
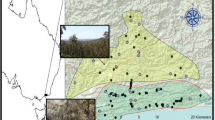
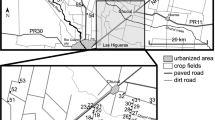
Anderson CS (2004) Effects of forest fragmentation on the abundance, distribution, and population genetic structure of white-footed mice (Peromyscus leucopus). Dissertation, Miami University, Oxford, Ohio, USA
Anderson CS, Meikle DB (2006) Annual changes in structural complexity of understory vegetation and relative abundance of Peromyscus leucopus in fragmented habitats. Acta Theriol 51:43–51
Anderson CS, Cady AB, Meikle DB (2003) Effects of vegetation structure and edge habitat on the density and distribution of white-footed mice (Peromyscus leucopus) in small and large forest patches. Can J Zool 81:897–904
Andren H (1994) Effects of habitat fragmentation on birds and mammals in landscapes with different proportions of suitable habitat: a review. Oikos 71:355–366
Andren H, Angelstam P (1988) Elevated predation rates as an edge effect in habitat islands: experimental evidence. Ecology 69:544–547
Austin JD, Lougheed SC, Boag PT (2004) Controlling for the effects of history and nonequilibrium conditions in gene flow estimates in northern bullfrog (Rana catesbeiana) populations. Genetics 168:1491–1506
Balloux F (2001) EASYPOP (version 1.7): a computer program for the simulation of population genetics. J Hered 92:301–302
Beerli P (2002) Migrate (version 1.6)—documentation and program. Available on the site http://evolution.genetics.washington.edu/larmarc.html
Beerli P (2004) Effect of unsampled populations on the estimation of population sizes and migration rates between sampled populations. Mol Ecol 13:827–836
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98:4563–4568
Berry O, Tocher MD, Sarre SD (2004) Can assignment tests measure dispersal? Mol Ecol 13:551–561
Bonin A, Bellemain E, Bronken Eidesen P et al (2004) How to track and assess genotyping errors in population genetic studies. Mol Ecol 13:3261–3273
Bowers MS, Dooley JL (1999) A controlled, hierarchical study of habitat fragmentation: responses at the individual patch and landscape scale. Landscape Ecol 14:381–389
Bowers MA, Matter SF (1997) Landscape ecology of mammals: relationships between density and patch size. J Mammal 78:999–1013
Bowman J, Cappuccino N, Fahrig L (2002) Patch size and population density: the effect of immigration behavior. Conserv Ecol 6:9–15
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–455
Brown AL, Litvaitis JA (1995) Habitat features associated with predation of New England cottontails: what scale is appropriate? Can J Zool 73:1005–1011
Bruseo JA, Vessey SH, Graham JS (1999) Discrimination between Peromyscus leucopus noveboracensis and Peromyscus maniculatus nubiterrae in the field. Acta Theriol 44:151–160
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Chirhart SE, Honeycutt RL, Greenbaum IF (2000) Microsatellite markers for the deer mouse Peromyscus maniculatus. Mol Ecol 9:1669–1671
Connor EF, Courtney AC, Yoder JM (2000) Individuals-area relationship: the relationship between animal population density and area. Ecology 81:734–748
Cornuet JM, Piry S, Luikart G et al (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000
Cummings JR, Vessey SH (1994) Agricultural influences on movement patterns of white-footed mice (Peromyscus leucopus). Am Midl Nat 132:209–218
Diaz M, Santos T, Telleria JL (1999) Effects of forest fragmentation on the winter body condition and population parameters of a habitat generalist, the wood mouse Apodemus sylvaticus: a test of hypotheses. Acta Oecol 20:39–49
Diffendorfer JE, Gaines MS, Holt RD (1995) Habitat fragmentation and movements of three small mammals (Sigmodon, Microtus, and Peromyscus). Ecology 76:827–839
Doonan TJ, Slade NA (1995) Effects of supplemental food on population dynamics of cotton rats, Sigmodon hispidus. Ecology 76:814–826
Duquette LS, Millar JS (1995) The effect of supplemental food on life-history traits and demography of a tropical mouse Peromyscus mexicanus. J Anim Ecol 64:348–360
Gibbs HL, Dawson RJG, Hobson KA (2000) Limited differentiation in microsatellite DNA variation among northern populations of the yellow warbler: evidence for male-biased gene flow? Mol Ecol 9:2137–2147
Gilbert BS, Krebs CJ (1981) Effects of extra food on Peromyscus and Clethrionomys populations in the Southern Yukon Canada. Oecologia 51:326–331
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available on the site http://www2.unil.ch/popgen/softwares/fstat.htm
Griffith DM, Wharton EH (1994) Ohio’s forests, looking good - but. Ohio Woodlands 31:20–21
Hanski I, Simberloff D (1997) The metapopulation approach, its history, conceptual domain, and application to conservation. In: Hanski I, Gilpin ME (eds) Metapopulation biology: ecology, genetics, and evolution. Academic Press, New York
Hoffman JI, Amos W (2005) Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol Ecol 14:599–612
Ishibashi Y, Saitoh T, Abe S et al (1996) Null microsatellite alleles due to nucleotide sequence variation in the grey-sided vole Clethrionomys rufocanus. Mol Ecol 5:589–590
Kaufman DW, Kaufman GA, Finck EJ (1995) Temporal variation in abundance of Peromyscus leucopus in wooded habitats of eastern Kansas. Am Midl Nat 133:7–17
Kinnison MT, Bentzen P, Unwin MJ et al (2002) Reconstructing recent divergence: evaluating nonequilibrium population structure in New Zealand Chinook salmon. Mol Ecol 11:739–754
Koenig WD, Van Vuren D, Hooge PN (1996) Detectability, philopatry, and the distribution of dispersal distances in vertebrates. Trends Ecol Evol 11:514–517
Krohne DT, Burgin AB (1990) Demographic heterogeneity in a population of Peromyscus leucopus. Oecologia 82:97–101
Krohne DT, Hoch GA (1999) Demography of Peromyscus leucopus populations on habitat patches: the role of dispersal. Can J Zool 77:1247–1253
Lackey JA (1978) Geographic variation in habitat use by the white-footed mouse, Peromyscus leucopus. Am Midl Nat 100:171–177
Lewellen RH, Vessey SH (1998) Modeling biotic and biotic influences on population size in small mammals. Oecologia 113:210–218
Lima SL, Dill LM (1990) Behavioral decisions made under risk of predation: a review and prospectus. Can J Zool 68:619–640
Linzey AV (1989) Response of the white-footed mouse (Peromyscus leucopus) to the transition between disturbed and undisturbed habitats. Can J Zool 67:505–512
M’Closkey RT (1975) Habitat dimension of white-footed mice, Peromyscus leucopus. Am Midl Nat 93:158–167
Maier TJ (2002) Long-distance movements by female white-footed Mice, Peromyscus leucopus , in extensive mixed-wood forest. Can Field Nat 116:108–111
Manel S, Berthier P, Luikart G (2002) Detecting wildlife poaching: identifying the origin of individuals with Bayesian assignment tests and multilocus genotypes. Cons Biol 16:650–659
Manier MK, Arnold SJ (2005) Population genetic analysis identifies source-sink dynamics for two sympatric garter snake species (Thamnophis elegans and Thamnophis sirtalis). Mol Ecol 14:3965–3976
Manson RH, Ostfeld RS, Canham CD (1999) Responses of a small mammal community to heterogeneity along forest-old-field edges. Landscape Ecol 14:355–367
Martell AM (1983) Demography of southern red-backed voles Clethrionomys gapperi and deer mice Peromyscus maniculatus after logging in north central Ontario Canada. Can J Zool 61:958–969
Matter SF (1997) Population density and area: the role of within and between patch processes. Oecologia 110:533–538
Middletown J, Merriam G (1981) Woodland mice in a farmland mosaic. J Appl Ecol 18:703–710
Mossman CA, Waser PM (1999) Genetic detection of sex-biased dispersal. Mol Ecol 8:1063–1067
Mossman CA, Waser PM (2001) Effects of habitat fragmentation on population genetic structure in the white-footed mouse (Peromyscus leucopus). Can J Zool 79:285–295
Narum SR (2006) Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv Genet 7:783–787
Nupp TE, Swihart RK (1996) Effect of forest patch area on population attributes of white-footed mice (Peromyscus leucopus) in fragmented landscapes. Can J Zool 74:467–472
Nupp TE, Swihart RK (1998) Effects of forest fragmentation on population attributes of white-footed mice and eastern chipmunks. J Mammal 79:1234–1243
Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Paetkau D, Strobeck C (1995) The molecular basis and evolutionary history of a microsatellite null allele in bears. Mol Ecol 4:519–520
Pearse DE, Crandall KA (2004) Beyond F ST: analysis of population genetic data for conservation. Conserv Genet 5:585–602
Pemberton JM, Slate J, Bancroft DR, Barrett JA (1995) Non-amplifying alleles at microsatellite loci: a caution for parentage and population studies. Mol Ecol 4:249–252
Piry S, Alapetite A, Cornuet JM et al (2004) GENECLASS2: a software for genetic assignment and first-generation migration detection. J Hered 95:536–539
Pulliam HR (1988) Sources, sinks, and population regulation. Am Nat 132:652–661
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201
Raymond M, Rousset F (1995) Population genetics software for exact test and eumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rintamaa DL, Mazur PA, Vessey SH (1976) Reproduction during two annual cycles in a population of Peromyscus leucopus noveborecensis. J Mammal 57:593–595
Roach JL, Stapp P, Van Horne B et al (2001) Genetic structure of a metapopulation of black-tailed prairie dogs. J Mammal 82:946–959
Root RB (1973) Organization of a plant-arthropod association in simple and diverse habitats: the fauna of collards, Brassica oleracea. Ecol Monogr 43:95–124
Saunders DA, Hobbs RJ, Margules CR (1991) Biological consequences of ecosystem fragmentation: a review. Conserv Biol 5:18–32
Schmid-Holmes S, Drickamer LC (2001) Impact of forest patch characteristics on small mammal communities: a multivariate approach. Biol Conserv 99:293–305
Schmidt CA (1999) Variation and congruence of microsatellite markers for Peromyscus leucopus. J Mammal 80:522–529
Skupski MP (1995) Population ecology of the western harvest mouse, Reithrodontomys megalotis: a long-term perspective. J Mammal 76:358–367
Slade NA, Blair SM (2000) An empirical test of using counts of individuals captured as indices of population size. J Mammal 81:1035–1045
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Slatkin M, Barton NH (1989) Methods for estimating gene flow. Evolution 43:1349–1368
Stapp P, Polis GA (2003) Marine resources subsidize insular rodent populations in the Gulf of California, Mexico. Oecologia 134:496–504
Szacki J (1999) Spatially structured populations: how much do they match the classic metapopulation concept. Landscape Ecol 14:369–379
Waser P, Strobeck C (1998) Genetic signatures of interpopulation dispersal. Trends Ecol Evol 13:43–44
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: F ST not equal 1/(4Nm +1). Heredity 82:117–125
Wilder S, Meikle DB (2005) Reproduction, foraging, and the negative density-area relationship of a generalist rodent. Oecologia 144:391–398
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wolf M, Batzli G (2004) Forest edge—high or low quality habitat for white-footed mice (Peromyscus leucopus)? Ecology 85:756–769
Wolff JO, Lundy KI, Baccus R (1988) Dispersal, inbreeding avoidance, and reproductive success in white-footed mice. Anim Behav 36:456–465
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420
Yahner RH (1988) Changes in wildlife communities near edges. Conserv Biol 2:333–339
Yahner RH (1992) Dynamics of a small mammal community in a fragmented forest. Am Midl Nat 127:381–391
Yunger JA (2002) Response of two low-density populations of Peromyscus leucopus to increased food availability. J Mammal 83:267–279
Acknowledgements
We thank the landowners and farmers who allowed us to use their land for study sites, in addition to Miami University’s Ecology Research Center, Miami University’s Natural Areas Committee, and the Ohio Department of Natural Resources. A. Abtahi, A. Lohrey, D. Miller, M. Thobe, and especially F. Madore provided valuable assistance in the field and laboratory. We thank D. Claussen, T. Crist, L. Gibbs, B. Keane, K. Mylecraine, R. Schaefer, and other anonymous reviewers for comments that greatly improved this manuscript. We also thank I. Greenbaum, C. Mossman, C. Van Oosterhout, and J. Roach for their insightful suggestions concerning this project. C. Wood in the Center for Bioinformatics and Functional Genomics at Miami University and S. Corey and J. Diaz in L. Gibbs’ laboratory at The Ohio State University graciously provided advice and assistance with genetic techniques and analyses. This work was supported with funding from the Graduate School and Department of Zoology at Miami University. Research on live animals was performed in a humane manner and was approved by the Institutional Animal Care and Use Committee (IACUC) at Miami University (protocol #522).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
Appendices
Rights and permissions
About this article
Cite this article
Anderson, C.S., Meikle, D.B. Genetic estimates of immigration and emigration rates in relation to population density and forest patch area in Peromyscus leucopus . Conserv Genet 11, 1593–1605 (2010). https://doi.org/10.1007/s10592-009-0033-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-0033-8