Abstract
Metastatic cells exhibit an extraordinary phenotypic plasticity, not only in adapting to unfamiliar microenvironments but also in surviving aggressive treatments and immune responses. A major source of phenotypic variability is alternative splicing (AS) of the pre-messenger RNA. This process is catalyzed by one of the most complex pieces of cellular molecular regulatory events, the spliceosome, which is composed of ribonucleoproteins and polypeptides termed spliceosome factors. With strong evidence indicating that AS affects nearly all genes encoded by the human genome, aberrant AS programs have a significant impact on cancer cell development and progression. In this review, we present insights about the genomic and epigenomic factors affecting AS, summarize the most recent findings linking aberrant AS to metastatic progression, and highlight potential prognostic and therapeutic applications.





Similar content being viewed by others
References
Feinberg AP, Koldobskiy MA, Gondor A (2016) Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17(5):284–299
Pujadas E, Feinberg AP (2012) Regulated noise in the epigenetic landscape of development and disease. Cell 148(6):1123–1131
Wang ET et al (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456(7221):470–476
Ghigna C et al (2010) Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol 7(4):495–503
Tang JY et al (2013) Alternative splicing for diseases, cancers, drugs, and databases. Sci World J 2013:703568
Koo T, Wood MJ (2013) Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum Gene Ther 24(5):479–488
Faravelli I et al (2015) Spinal muscular atrophy-recent therapeutic advances for an old challenge. Nat Rev Neurol 11(6):351–359
Bonnal S, Vigevani L, Valcarcel J (2012) The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 11(11):847–859
Matera AG, Wang Z (2014) A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15(2):108–121
Park E et al (2018) The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet 102(1):11–26
Zhang J, Manley JL (2013) Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov 3(11):1228–1237
Anczukow O, Krainer AR (2016) Splicing-factor alterations in cancers. RNA 22(9):1285–1301
DeBoever C et al (2015) Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput Biol 11(3):e1004105
Alsafadi S et al (2016) Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun 7:10615
Lee SC, Abdel-Wahab O (2016) Therapeutic targeting of splicing in cancer. Nat Med 22(9):976–986
Kim E et al (2015) SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27(5):617–630
Okeyo-Owuor T et al (2015) U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 29(4):909–917
Shirai CL et al (2015) Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell 27(5):631–643
Zhang J et al (2015) Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci USA 112(34):4726–4734
Ilagan JO et al (2015) U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res 25(1):14–26
Imielinski M et al (2012) Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150(6):1107–1120
Lev Maor G, Yearim A, Ast G (2015) The alternative role of DNA methylation in splicing regulation. Trends Genet 31(5):274–280
Zhou HL et al (2014) Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res 42(2):701–713
Luco RF et al (2010) Regulation of alternative splicing by histone modifications. Science 327(5968):996–1000
Wong JJ et al (2014) Epigenetic modifications of splicing factor genes in myelodysplastic syndromes and acute myeloid leukemia. Cancer Sci 105(11):1457–1463
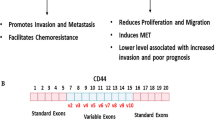
Marzese DM et al (2015) Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res 28(1):82–93
Maunakea AK et al (2013) Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res 23(11):1256–1269
Saint-Andre V et al (2011) Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol 18(3):337–344
Warzecha CC, Carstens RP (2012) Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol 22(5–6):417–427
Oltean S, Bates DO (2014) Hallmarks of alternative splicing in cancer. Oncogene 33(46):5311–5318
David CJ, Manley JL (2010) Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24(21):2343–2364
Zhang P et al (2013) Exon 4 deletion variant of epidermal growth factor receptor enhances invasiveness and cisplatin resistance in epithelial ovarian cancer. Carcinogenesis 34(11):2639–2646
Solomon H, Sharon M, Rotter V (2014) Modulation of alternative splicing contributes to cancer development: focusing on p53 isoforms, p53beta and p53gamma. Cell Death Differ 21(9):1347–1349
Avery-Kiejda KA et al (2014) The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis 35(3):586–596
Jin W et al (2000) Fibroblast growth factor receptor-1 alpha-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res 60(5):1221–1224
He X et al (2014) Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis 3:e84
He X et al (2007) Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene 26(34):4961–4968
Cheung HC et al (2009) Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain 132(Pt 8):2277–2288
Silipo M, Gautrey H, Tyson-Capper A (2015) Deregulation of splicing factors and breast cancer development. J Mol Cell Biol 7(5):388–401
Pradella D et al (2017) EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 16(1):8
Warzecha CC et al (2010) An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 29(19):3286–3300
Anczukow O et al (2015) SRSF1-regulated alternative splicing in breast cancer. Mol Cell 60(1):105–117
Warzecha CC et al (2009) ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33(5):591–601
Das S, Krainer AR (2014) Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 12(9):1195–1204
Ghigna C et al (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20(6):881–890
Karni R et al (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14(3):185–193
Ben-Hur V et al (2013) S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep 3(1):103–115
Ishii H et al (2014) Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 289(40):27386–27399
Yae T et al (2012) Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun 3:883
Chen L et al (2017) Snail driving alternative splicing of CD44 by ESRP1 enhances invasion and migration in epithelial ovarian cancer. Cell Physiol Biochem 43(6):2489–2504
The Cancer Genome Atlas Network (2015) Genomic classification of cutaneous melanoma. Cell 161(7):1681–1696
Ryan MC et al (2012) SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 28(18):2385–2387
Maatz H et al (2014) RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest 124(8):3419–3430
Darman RB et al (2015) Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep 13(5):1033–1045
Harbour JW et al (2013) Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet 45(2):133–135
Biankin AV et al (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491(7424):399–405
Martin M et al (2013) Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 45(8):933–936
Quesada V et al (2011) Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44(1):47–52
Kaida D et al (2007) Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3(9):576–583
Kotake Y et al (2007) Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3(9):570–575
Hasegawa M et al (2011) Identification of SAP155 as the target of GEX1A (herboxidiene), an antitumor natural product. ACS Chem Biol 6(3):229–233
Graveley BR, Maniatis T (1998) Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell 1(5):765–771
Zhou Z, Fu XD (2013) Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122(3):191–207
Twyffels L, Gueydan C, Kruys V (2011) Shuttling SR proteins: more than splicing factors. FEBS J 278(18):3246–3255
Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417(1):15–27
Araki S et al (2015) Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing. PLoS ONE 10(1):e0116929
Gammons MV et al (2014) Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma. Br J Cancer 111(3):477–485
Ohe K, Hagiwara M (2015) Modulation of alternative splicing with chemical compounds in new therapeutics for human diseases. ACS Chem Biol 10(4):914–924
Kole R, Krainer AR, Altman S (2012) RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11(2):125–140
Zammarchi F et al (2011) Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci USA 108(43):17779–17784
Dewaele M et al (2016) Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 126(1):68–84
Castanotto D, Stein CA (2014) Antisense oligonucleotides in cancer. Curr Opin Oncol 26(6):584–589
Moreno PM, Pego AP (2014) Therapeutic antisense oligonucleotides against cancer: hurdling to the clinic. Front Chem 2:87
Peng Y-Q et al (2017) Applications of CRISPR/Cas9 in retinal degenerative diseases. Int J Ophthalmol 10(4):646–651
Bengtsson NE et al (2017) Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 8:14454
Nguyen TH, Anegon I (2016) Successful correction of hemophilia by CRISPR/Cas9 genome editing in vivo: delivery vector and immune responses are the key to success. EMBO Mol Med 8(5):439–441
Feng Y et al (2015) Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system. J Orthop Res 33(2):199–207
Liao Y et al (2017) Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget 8(18):30276–30287
Sanchez-Rivera FJ, Jacks T (2015) Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer 15(7):387–395
Torres-Ruiz R, Rodriguez-Perales S (2015) CRISPR-Cas9: a revolutionary tool for cancer modelling. Int J Mol Sci 16(9):22151–22168
Zhou Q et al (2015) A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res 75(10):1949–1958
Bustos M et al (2018) Abstract P1-05-02: CRISPR/Cas9-guided editing of spliceosome factors enhances major histocompatibility complex proteins in triple-negative breast cancer. Can Res 78(4 Supplement):P1-05-02
Eskens FA et al (2013) Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res 19(22):6296–6304
Hong DS et al (2014) A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs 32(3):436–444
Seiler M et al (2018) H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat Med 24(4):497–504
Bedikian AY et al (2006) Bcl-2 antisense (oblimersen sodium) plus dacarbazine in patients with advanced melanoma: the Oblimersen Melanoma study group. J Clin Oncol 24(29):4738–4745
O’Brien S et al (2007) Randomized phase III trial of fludarabine plus cyclophosphamide with or without oblimersen sodium (Bcl-2 antisense) in patients with relapsed or refractory chronic lymphocytic leukemia. J Clin Oncol 25(9):1114–1120
Chanan-Khan AA et al (2009) Phase III randomised study of dexamethasone with or without oblimersen sodium for patients with advanced multiple myeloma. Leuk Lymphoma 50(4):559–565
Chi KN et al (2010) Randomized phase II study of docetaxel and prednisone with or without OGX-011 in patients with metastatic castration-resistant prostate cancer. J Clin Oncol 28(27):4247–4254
Beer TM et al (2017) Custirsen (OGX-011) combined with cabazitaxel and prednisone versus cabazitaxel and prednisone alone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel (AFFINITY): a randomised, open-label, international, phase 3 trial. Lancet Oncol 18(11):1532–1542
Chi KN et al (2017) Custirsen in combination with docetaxel and prednisone for patients with metastatic castration-resistant prostate cancer (SYNERGY trial): a phase 3, multicentre, open-label, randomised trial. Lancet Oncol 18(4):473–485
Acknowledgements
This work was supported by the National Cancer Institute, National Institutes of Health (#R01CA167967); the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation; the AVON Foundation Breast Cancer Crusade (#02-2015-061); the Associates for Breast and Prostate Cancer Studies (ABCs) award; and the Fashion Footwear Association of New York (FFANY) foundation award.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Marzese, D.M., Manughian-Peter, A.O., Orozco, J.I.J. et al. Alternative splicing and cancer metastasis: prognostic and therapeutic applications. Clin Exp Metastasis 35, 393–402 (2018). https://doi.org/10.1007/s10585-018-9905-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-018-9905-y