Abstract
The basal lineage of monotremes features an extraordinarily complex sex chromosome system which has provided novel insights into the evolution of mammalian sex chromosomes. Recently, sequence information from autosomes, X chromosomes, and XY-shared pseudoautosomal regions has become available. However, no gene has so far been described on any of the Y chromosome-specific regions. We analyzed sequences derived from Y-specific BAC clones to identify genes with potentially male-specific function. Here, we report the identification and characterization of the mediator complex protein gametologs on platypus Y5 (Crspy). We also identified the X-chromosomal copy which unexpectedly maps to X1 (Crspx). Sequence comparison shows extensive divergence between the X and Y copy, but we found no significant positive selection on either gametolog. Expression analysis shows widespread expression of Crspx. Crspy is expressed exclusively in males with particularly strong expression in testis and kidney. Reporter gene assays to investigate whether Crspx/y can act on the recently discovered mouse Sox9 testis-specific enhancer element did reveal a modest effect together with mouse Sox9 + Sf1, but showed overall no significant upregulation of the reporter gene. This is the first report of a differentiated functional male-specific gene on platypus Y chromosomes, providing new insights into sex chromosome evolution and a candidate gene for male-specific function in monotremes.
Similar content being viewed by others
Introduction
Heteromorphic sex chromosomes have evolved in different animal and plant lineages. In some lineages, sex chromosomes have been conserved (for example, the XY sex chromosomes in therian mammals and the ZW in birds), while different sex chromosomes have evolved in closely related insect, fish, and reptile species (except birds) (Ming et al. 2011; Kaiser and Bachtrog 2010; Marshall Graves 2008). Despite utilizing different sex chromosomes, the differentiation of male- or female-specific Y and W chromosomes due to lack of recombination is a common theme in sex chromosomes evolution, and leads to similar patterns of sequence differentiation on X/Y or Z/W chromosomes (Charlesworth et al. 2005; Ellegren 2011). The generally accepted model of sex chromosome evolution is that accumulation of sexually antagonist genes in proximity of a sex-determining locus provides a selective advantage for lack of recombination between sex chromosomes (Charlesworth 1991). While the X and Z chromosomes will undergo recombination in the homogametic sex, the Y and W chromosomes will accumulate mutations. Deleterious mutations can accumulate faster because of the fixed heterozygosity of Y and W chromosomes in the heterogametic sex and other evolutionary forces such as Muller’s ratchet and genetic hitchhiking (Felsenstein 1974; Rice 1987; Muller 1918). On the level of single genes, it has been suggested that accumulation of deleterious mutations is due to relaxed purifying selection in combination with genetic drift, enhanced by low effective population size. Alternatively, genetic hitchhiking would allow mutations to be fixed in particular under strong positive selection. Together, these models predict increased levels of nonsynonymous (dN) to synonomyous (dS) substitutions on heteromorphic sex chromosomes (Berlin and Ellegren 2006; Sandstedt and Tucker 2005; Wyckoff et al. 2002).
Several mechanisms have been proposed to slow this inevitable decay of one of the sex chromosomes. This includes palindromic sequence organization facilitating gene conversion (Rozen et al. 2003; Skaletsky et al. 2003) and occasional recombination events including X- and Y-specific regions have been proposed recently (Stock et al. 2011). Differentiation of genes as a result of sex chromosome evolution presents an opportunity for subfunctionalization and neofunctionalization of the gene on the sex-specific chromosome (Y or W chromosome). This can occur via mutation in the non-recombining part and the frequent duplication events that often result in multiple copies of genes on Y chromosomes (Wilson and Makova 2009). Signatures of subfunctionalization and neofunctionalization are change in the expression pattern and sequence divergence. Such a pattern of gene evolution has been shown for genes that have retroposed from autosomes to Y chromosomes (e.g., CDYL/CDY in primates and DMRT1/DMY in medaka) or gametologs that diverged as a result of recombination suppression (Wilson and Makova 2009; Matsuda et al. 2007; Dorus et al. 2003).
In addition to the common XX/XY and ZZ/ZW sex chromosomes systems, some species feature more than two sex chromosomes. These can arise by interchromosomal rearrangements between an autosome and a sex chromosome. Usually, these autosome–sex–chromosome rearrangements are thought to be deleterious as they interfere with sex chromosome-specific epigenetic changes like X inactivation or meiotic sex chromosome inactivation (Ashley 2002). Despite this, X1X2Y and rarely XY1Y2 sex chromosome system have been described in various mammalian lineages and are frequently found in fish and insects (Gruetzner et al. 2006; Cioffi et al. 2011a, b; Galian et al. 2007; Rowell 1987). In most cases, these systems are thought to have evolved independently.
The multiple sex chromosome system in monotreme mammals defies most of the rules that seem to apply to other sex chromosome multiples (Gruetzner et al. 2006). Both platypus (Ornithorhynchus anatinus) and echidna (shortbeaked echidna, Tachyglossus aculeatus and three species of long-beaked echidna, Zaglossus), constituting the extant monotreme lineage, feature multiple sex chromosome system including ten and nine sex chromosomes, respectively (Murtagh 1977; Grutzner et al. 2004; Rens et al. 2004, 2007; Bick et al. 1973). Eight sex chromosomes share homology between platypus and echidna (Tachyglossus aculeatus) showing that this system evolved early in the monotreme lineage (Rens et al. 2007). Additional autosome–sex–chromosome translocations in the echidna and platypus lineage occurred after their divergence over 25 Million years ago (Rens et al. 2007; Dohm et al. 2007; Warren et al. 2008). There are two fundamentally different scenarios to explain the evolution of this complex system (Gruetzner et al. 2006). The difference between the platypus and echidna sex chromosomes, and in particular, the scattering of homology to the chicken Z chromosome on X5, X3, X2, and X1 supports a sequential sex chromosome autosome translocation model (Grutzner et al. 2004; Veyrunes et al. 2008). In addition, homology to a number of autosomes has been established but notably no homology to the human X chromosomes has been found on any of the sex chromosomes. Instead, most of the platypus orthologs of genes on the human or mouse X have been mapped to platypus chromosome 6 (Veyrunes et al. 2008; Rens et al. 2007).
Complexity and evolutionary ancestry of monotreme sex chromosomes can provide valuable insights into a number of aspects of sex chromosome biology. The main knowledge gap regarding monotreme sex chromosomes at present is the absence of information about the Y-specific parts of the four Y chromosomes in echidna and five Y chromosomes in platypus. This is particularly relevant as the sex determination mechanism in these species is not yet understood. In most other mammals, the Y-borne Sry gene acts via the testis-specific enhancer element to upregulate Sox9 during early male development (Sekido and Lovell-Badge 2008). Although the enhancer element is present in platypus (Bagheri-Fam et al. 2010), monotremes lack the Sry gene (Grutzner et al. 2004) and it is unclear how sex determination is triggered and whether it is mediated through Sox9 upregulation.
The platypus genome project sequenced the genome of a female platypus providing information about the gene content of mainly the larger X1 and X5 with some genes assigned to X2 and X3, but no sequence information has been identified on the very small X4 (Warren et al. 2008). The genome project therefore revealed information about X-specific and XY-shared regions but not for Y-specific regions. Monotremes are not unusual in this, and comprehensive sequence information has been missing from many species including humans where the sequence of the Y chromosome was determined several years after the initial sequencing of the human genome. Only recently, sequence information has been published for other mammalian species (Hughes et al. 2006; Skaletsky et al. 2003; Murphy et al. 2006). Together, these illustrate the need to obtain sequence information that will provide insights into the process of Y chromosome differentiation and reveal genes potentially involved in sex determination in monotremes. Because of a generally poor physical map, and complete absence of clones mapped to Y chromosomes, we used Y chromosome paints to isolate and sequence Y-specific bacterial artificial chromosomes (BAC) clones (Kortschak et al. 2009; Grutzner et al. 2004). Sequencing of those Y-specific clones has already revealed insights about the repeat and gene content of platypus Y chromosomes (Kortschak et al. 2009). In this study, we present the identification and analysis of a mediator complex subunit gene with differentiated X and Y copies in platypus.
Material and methods
RNA extraction
Adult male (liver, muscle, kidney, testis, brain) and female (liver, muscle, kidney, brain, active ovary) platypus tissues were used to isolate DNA and RNA using standard protocols. Total RNA was extracted from −80°C frozen tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA was resuspended in nuclease free water and stored at −80°C. To remove genomic DNA, 1 μg of RNA was treated with DNase I (New England Biolabs, USA), precipitated and re-suspended in RNase-free H2O.
cDNA synthesis
Samples of 1 μg of RNA were used to obtain cDNA from each tissue using a SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA). Briefly, RNA was incubated with 1 μl of 50 μM oligo(dT) and 1 μl of 10 mM dNTP for 5 min at 65°C. After incubation, 2 μl of 10× RT buffer, 4 μl of 25 mM MgCl2, 2 μl of dithiothreitol (DTT, 0.1 M), 1 μl of RNaseOUT™ (40 U/μl), and 1 μl SuperScript III RT (200 U/μl) were added and incubated for 50 min at 50°C. The reaction was terminated at 85°C for 5 min. To increase the sensitivity of the PCR reactions, 1 μl of RNase H was added to each tube and incubated further for 20 min at 37°C. The cDNA synthesis reactions were stored at −20°C.
Gene expression analysis
RT-PCR was optimized and performed to determine the level of mRNA expression in 6 platypus tissues. The PCR was performed in 25 μl volume containing 0.5 μl of cDNA from each tissue, 2.5 μl of 10× PCR reaction buffer with MgCl2, 1 μl of 10 mM dNTP solution, 0.5 μl of each primer (20 pmol/μl), and 0.125 μl (1 U/μl) of Roche Taq DNA polymerase (Mannheim, Germany). The temperature conditions for all PCRs were as follows: initial denaturation at 95°C for 2 min, 5 cycles of denaturation at 95°C for 30 s, annealing at 70°C for 4 min, extension at 72°C for 2 min and 25 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 45 s, extension at 72°C for 2 min, and 5 min for the final extension. The PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide.
Platypus Crspx and Crspy primers were designed using NCBI primer-blast online program at http://www.ncbi.nlm.nih.gov/tools. Primers are shown 5′–3′ as follows: Crspx for-ACCCGTCGCCCAGAATGAAGC, Crspx rev-GGTCGGGAGCGGAGAGGAGTT; Crspy for-ACCAGTAAATGCTGTGAAACCTC, Crspy rev-TTCTTTTTATTGGCTGGTTCTGA. Specificity of each primer set was confirmed by cloning of the PCR products into pGEM-T Easy vector (Promega, Madison, USA) and subsequent sequencing. The PCR products were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit. To amplify distinguishable products in genomic DNA and cDNA and to avoid DNA contamination, primers were designed to span an intron. Sequence similarities to known sequences were identified using NCBI BLAST analysis. For statistical significance, all RT-PCR experiments for each primer pair were performed in triplicate and repeated three times on separate biological samples. The platypus Crspx and Crspy positive BAC clones were confirmed by PCR using abovementioned primers. Platypus beta-Actin primers bAfor- GCCCATCTACGAAGGTTACGC and bArev-AAGGTCGTTTCGTGGATACCAC were used as internal and loading controls. For all samples, the positive PCR products for each gene were confirmed by sequencing and compared with the platypus genome database (data not shown).
Phylogenetic analysis
Sequences of the following species were included in the analysis: zebrafish (Danio rerio) AY034615.1, frog (Xenopus laevis) NM_001090429.1, chicken (Gallus gallus) XM_001234570.1, opossum (Monodelphis domestica) XM_001363095.1, platypus (Ornithorhynchus anatinus), and human (Homo sapiens) AF104253.1 (see Supplementary data)
cDNA sequences for platypus Crspx and Crspy were manually curated. Crspx sequence was generated from Contig8610 and Contig6275 to maximize similarity with other Crsp7 genes considering genomic features such as predicted splice sites, and the Crspy sequence used in this analysis is a chimera of three distinct Crspy loci. Corrections of the sequences in the database can be accessed on the NCBI gene database. GenBank accession number for platypus Crspy is JN815314. Sequences were aligned using MUSCLE (Edgar 2004) with the MEGA package (Tamura et al. 2011) using the following parameters: codon alignment, gap open −2.9, gap extend 0, hydrophobicity multiplier 1.2, max iterations 8, UPGMA clustering, min diag length 24, and standard genetic code (i.e., default options). MrBayes (Huelsenbeck and Ronquist 2001)—version 3.1.2 was used to reconstruct the tree with the following parameters: 106 iterations using GTR + gamma and proportion of invariant sites. PyCogent (Knight et al. 2007)—version 1.5 was used to determine dN/dS values for branches.
BAC clone isolation
Platypus Y5 chromosome BAC (OABb-285J12) was identified and sequenced as previously described (Kortschak et al. 2009; Grutzner et al. 2004). The Crsp7 gene sequence (ENSOANT00000021231, http://www.ensembl.org) was used to identify a BAC clone CH236-456I13 containing the platypus Crspx gene (http://www.ncbi.nlm.nih.gov/Traces and http://genome.wustl.edu/tools/blast). The platypus BAC clones (OABb-285J12 and CH236-456I13) were obtained from the CUGI BAC/EST resource center (http://www.genome.clemson.edu) and CHORI-236 BAC clone (http://bacpac.chori.org).
Illumina sequencing
Illumina libraries were prepared from cDNA subjected to fragmentation. Sample processing involving end repair, A-tailing, and ligation of Illumina single read adapters and was performed according to the recommendations by Illumina. We sequenced one lane each from the testis library and from the liver library on an Illumina GA I instrument, using a 36-cycle recipe. Basecalling was performed using Illumina pipeline v0.2.2.5
Physical mapping
Preparation of chromosomes
Mitotic metaphase chromosomes were generated from the established platypus fibroblast cell lines (Grutzner et al. 2004). Primary cultures were set up from toe-web tissue.
Fluorescence in situ hybridization of BAC clones
The chromosomal location of the Crspx and Crspy containing BAC clones (CH236-456I13 and OABb-285J12) was determined by fluorescence in situ hybridization of male platypus fibroblasts as described previously (Tsend-Ayush et al. 2009). Images were taken with a Zeiss AxioImager Z.1 epifluorescence microscope equipped with a CCD camera and Zeiss Axiovision software.
Reporter gene assay
Construction of platypus Crspx and Crspy expression plasmids: the 2.4-kb sequence of Crspy and the 1.6-kb sequence of Crspx were amplified using cDNAs derived from platypus testis by PCR with the sense primer Crspy-KpnI 5′-GGTACCACAATGACTGCGGA-3′, the antisense primer Crspy-ApaI 5′-GGGCCCTCAGTCTAGGCAGA-3′, the sense primer Crspx-AflII 5′-CTTAAGAACATGGTGGCAGT-3′ and the antisense primer Crspx-XbaI 5′-CAGTCTAGACAGACGTAAGG-3′. The resultant amplicons were agarose gel-purified and inserted into pGEM®-T Easy Vector (Promega, Madison, WI, USA) and further subcloned into pcDNA3.1(+) to form the Crspx and Crspy-expressing plasmids (pcDNA-Crspx, pcDNA-Crspy). The clones were confirmed by restriction digestion and sequencing.
Cell culture and transfections
HEK293T cells were grown in DMEM (Gibco) medium supplemented with 10% fetal bovine serum and L-glutamine in an atmosphere of 5% CO2. Transient transfections were conducted using Fugene6 (Roche) in accordance with the manufacturer’s instructions. Cells were seeded at a density of 0.5×105 cells per well in 12-well plates 24 h prior to transfection. Forty-eight hours post-transfection cell lysate was collected and luciferase reporter activity was measured according to the manufacturer’s instructions (Promega). Reporter activity was normalized to b-galactosidase as an internal control (Promega).
Results
Identification and characterization of the Crsp7 gametologs Crspx and Crspy in platypus
We obtained approximately 500 kb of sequence derived from Y-specific BAC clones (Kortschak et al. 2009). Sequence analysis revealed the presence of a gene with homology to the human mediator complex component 26 (Med26 or Crsp7). In the database Crsp7 (Crspx) maps to platypus genome SuperContig6275: 6,700–9,957 bp (www.ensembl.org, accessed September 2011), which has not been assigned to a specific chromosome (mapped to platypus X1 in this study, and hence termed Crspx). There are two platypus Crsp7 transcripts identified in the database. However, we obtained the full-length platypus Crspx cDNA from testis and kidney and sequencing of ten clones confirmed presence of only one transcript (ENSOANT00000021231).
The Y-specific 1,800-bp gene encodes a putative platypus Crspy protein of 599 amino acids with 57% identity to the predicted platypus Crspx protein and 53% identity to the human CRSP7 protein. Multiple alignments of the amino acid sequences derived from the Crsp7 family of genes from human, mouse, marsupial, and fish confirmed that there is moderate conservation across the entire protein, with the N-terminal transcription elongation factor (TfIIS-N) domain being the most conserved region (supplementary Fig. 1).
Phylogenetic analysis
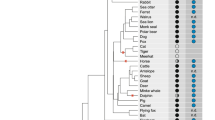
Species included in the analysis were zebrafish (D. rerio), frog (X. laevis), chicken (G. gallus), opossum (M. domestica), platypus (O. anatinus), and human (H. sapiens). Figure 1 shows a Bayesian reconstruction based on the cDNA sequence of these species (multiple alignment shown in supplementary Fig. 1). All nodes have a posterior probability of 1; the dichotomies contributing to the polytomy do not attain majority support. Surprisingly, platypus Crspy and Crspx are associated with different nodes in this reconstruction. We calculated the dN/dS changes to identify changes in selective pressure. The omega values obtained (see Fig. 1) did indicate relaxation of purifying selection in the case of platypus, a situation which may be explained by the existence of two distinct Crsp7 orthologs (gametologs) in this species allowing some diversification of function.
Phylogenetic reconstruction of the gene tree for a collection of Crsp7 orthologs and the platypus gametologs. Bayesian reconstruction based on the cDNA sequence of zebrafish (D. rerio), frog (X. laevis), chicken (G. gallus), opossum (M. domestica), platypus (O. anatinus), and human (H. sapiens). All nodes have a posterior probability of 1. Omega values (dN/dS) for the individual genes are depicted on the branches
Mapping of Crspx and Crspy in platypus
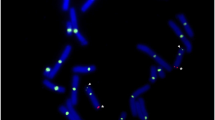
Fluorescence in situ hybridization was used to map the verified BAC clones onto platypus metaphase chromosome spreads. A BAC clone containing the platypus Crspy hybridized to the platypus Y-specific region of the minute Y5 chromosome (Fig. 2b). We used the available Crspx contig sequence to search the BAC sequence database (see Material and methods). A confirmed (PCR and sequencing) Crspx containing BAC clone hybridized unequivocally to the telomeric region of the long arm of platypus chromosome X1 and not to X5 as expected (Fig. 2a). The implications for understanding the evolution of the monotreme sex chromosome chain are discussed below.
Physical mapping of Crspx and Crspy in platypus. Fluorescence in situ hybridization of the Crspx containing BAC clone CH236-456I13 (labeled with spectrum orange (a)) and of the Crspy containing OABb-285J12 (labeled with spectrum green (b)) to male platypus metaphase chromosome spreads. Strong signals for Crspx were observed on the telomeric region of the long arm of platypus chromosome X1 and for Crspy on the smallest platypus chromosome Y5. The chromosomes are counterstained with DAPI. The inset shows a close up of the position of the small DAPI-weak platypus Y5. Scale bar is 5 μm
Expression analysis
RT-PCR was carried out to assess the transcriptional activity of the Crspx and Crspy genes in adult platypus tissues (brain, muscle, kidney, liver, testis, and ovary). The results demonstrated that the transcript of platypus Crspx was detected in all tested male and female tissues (Fig. 3). Crspy showed much higher expression in testis than kidney and low expression in liver, muscle, and brain. No Crspy expression was detected in female tissues tested (liver, kidney, brain, muscle, ovary, shown only for brain and ovary in Fig. 3).
Expression analysis of Crspx and Crspy in platypus. RT-PCR was performed to obtain the expression pattern of each gene in tissues from male and female adult platypus. The negative control is indicated as −ve. The platypus β-Actin gene was used as positive control. Right panel shows PCR on male and female genomic DNA showing male-specific bands for Crspy
Using Bowtie, we also determined the number of reads that mapped to Crspx and Crspy in two RNA-seq datasets derived from male platypus liver and testis samples (Langmeade et al. 2009). From the 6069551 liver-derived reads, we detected only six Crsp-aligning reads, all of which aligned with Crspx. While from the 7605063 testis-derived reads, 142 alignments were reported, 133 for Crspy and 9 for Crspx. This confirms our RT-PCR results of a uniform expression of Crspx in these tissues and a stronger expression of Crspy in testis. This expression difference between the Crspx and Crspy genes may indicate subfunctionalization of the male-specific Y copy of this gene in testis.
Reporter gene analysis of Crspx and Crspy
Monotremes lack the Sry gene which has recently been shown to activate Sox9 via a testis-specific enhancer element (Sekido and Lovell-Badge 2008). We wanted to test our hypothesis that Crspy would be efficient in activating the mouse TESCO (testis-specific enhancer core element) as a possible way of triggering male sex determination in platypus. To do this, platypus, Crspx and Crspy were cloned into an expression vector and transfected into cells containing the mouse TESCO enhancer linked to a luciferase reporter. Mouse genes included in the transfections which are potential partners were: Sf-1 which has been shown to bind the TESCO enhancer together with Sry in mouse, Sox3 which is the gametolog of Sry on the mouse X chromosomes (platypus has a Sox3 gene which resides on an autosome) and Sox9, which has been shown to efficiently auto upregulate via TESCO. Crspy and Crspx were co-transfected with those mouse genes and the relative luciferase activity and was measured and fold induction of the reported calculated (Fig. 4). As expected, Sox9 together with Sf-1 shows the highest induction of the reporter in this assay. Overall, both Crspx and Crspy do not cause a significantly increased induction with any of the mouse genes tested in this assay. However, we did observe a modest increase in particular of Crspx in combination with Sox9 and Sf-1 in this assay. From these experiments, it seems unlikely that Crspy acts as activator via TESCO of Sox9 in platypus.
Crspx and Crspy reporter gene assay. HEK293T cells were co-transfected with 100 ng of the TESCO-luciferase reporter in the presence of 300 ng of expression plasmids as indicated. The CMV-b-galactosidase reporter was used as internal control of transfection efficiency. Luciferase activity (a) was normalized to b-galactosidase activity (b) shows the fold induction of the reporter in this assay. Error bars indicate standard errors of the means for triplicate samples from three independent experiments
Discussion
Gene evolution on differentiated sex chromosomes is subject to a variety of evolutionary forces. This is due to the lack of recombination and is potentially accelerated by hitchhiking effects or sub-functionalization (and positive selection) of the male-specific copy of gametologs on the Y chromosomes. Monotreme sex chromosomes are a fascinating system in which to study Y chromosome differentiation. In addition, the sex-determining gene is still unknown. In order to obtain sequences from Y chromosomes, we have isolated and sequenced BAC clones from Y-specific regions and obtained initial sequences from some of those clones. Here, we report the identification and characterization of the differentiated X and Y copies of a mediator complex gene.
Evolution of Crspx and Crspy
The mediator complex is a large multi-protein complex which is required as transcriptional co-factor for gene-specific transcription factors. In this role, the mediator complex coordinates RNA polymerase II recruitment and activation. About 30 mediator complex genes have been identified in humans (Malik and Roeder 2010). Mediator genes are highly conserved in metazoan evolution. Mediator 26 is a subunit of the CRSP complex (co-factor required for SP1 activation) and maps to human chromosome 19p13.11 (Ryu et al. 1999). The transcription factor SP1 regulates a vast number of genes and SP1 binding sites can also been identified on a number of genes involved in sex determination in mammals including Sry (Desclozeaux et al. 1998). MED26 is part of only a small number of fractions that have been isolated from the 30 or so multi-protein complex. Recently the detailed mechanism of MED26 function has been resolved and a large number of target genes have been identified (Takahashi et al. 2011).
Interestingly, our identification of gametologs of mediator complex gene is not the first example of a Y copy of a gene from this group. In frog (Hyla arborea), orthologs of the Med15 gene are located on sex chromosomes. In this example, the X and Y copies of the Med15 gene have been highly conserved and differ only by a few frame shift-preserving indels in the glutamine-rich domain (Niculita-Hirzel et al. 2008). The differences between the platypus Crspx and Crspy genes are more substantial and could lead to significant changes in protein folding and function. Differentiation of sex-specific function may have been facilitated by the sex-specific existence and expression of Crspy.
There has been controversy over whether Y chromosome degradation is driven by random genetic drift in combination with relaxed purifying selection, or genetic hitchhiking where the deleterious mutation is fixed as a result of linkage to positively selected loci.
In the case of Crspx and Crspy, we calculated omega values substantially higher than for other branches in the analyzed clades (Fig. 1), suggesting relaxed purifying selection or possibly diversifying selection. A contributing factor maybe that effective population size in monotremes is small, increasing the effects of genetic drift possibly weakening purifying selection. In addition, monotremes are seasonal breeders with massively enlarged testis during breeding season. During sperm maturation, sperm form bundles, an approach which has been interpreted as result of fierce sperm competition (Djakiew and Jones 1982, 1983; Jones et al. 2004, 2007; Grutzner et al. 2008).
Our mapping of Crspy on Y5 and Crspx on X1 is surprising and the first example of homology between platypus X1 and Y5. This raises the question of when that differentiation of X and Y copy of this gene occurred in monotremes and how this may contribute to our understanding of the evolution of the sex chromosome complex.
Two different models have been proposed to explain the evolution of monotreme sex chromosomes (discussed in Gruetzner et al. 2006). The initial proposal that the multiple sex chromosome chain arose by recurrent translocations (Grutzner et al. 2004) was regarded as less parsimonious than a hybridization model which first results in a ring of translocation heterozyote chromosomes (Gruetzner et al. 2006; Ashley 2005). If the differentiation of the region containing Crspx and Crspy occurred more recently (i.e., before the divergence of platypus and echidna), it may have led a ring of ten chromosomes to open as a chain (supplementary Fig. 2).
If differentiation of the region containing Crspx and Crspy predates the first translocation event, this results in a chain and explains the location of Crspx on X1 and Crspy on Y5 after four X autosome translocations (Fig. 5). The scattering of homologies to the chicken Z chromosome extending from X5 to X1 was predicted by the translocation model in Gruetzner et al. 2006 and is supported by the disruption of the MHC region onto two sex chromosome pairs (Dohm et al. 2007), and importantly by the fact that different autosomes have been added to the sex chromosome complex in platypus and echidna independently (Rens et al. 2007). Given this evidence, Crspx/Crspy likely have differentiated on heteromorphic sex chromosomes before X autosome translocations led to this sex chromosome complex (Fig. 5).
Evolution of the platypus sex chromosome chain by recurrent reciprocal translocations. This model requires a differentiated ancestral sex chromosome pair. Four X-autosome reciprocal translocations (X-A translocation 1-4) will move Crspx from its original position on the ancestral X to the X1 and result in a sex chromosome chain of ten. The red bars indicate the translocation breakpoints. X–Y differentiation and Y chromosome degeneration leads to the reduction in size of the Y chromosomes observed in the platypus sex chromosome chain. An alternative model is given in supplementary Fig. 2
Crspy, a potential sex-determining gene in platypus?
Monotremes have no Sry gene, but the therian gametolog Sox3 is present on chromosome 6 (Wallis et al. 2007). Recently, the crucial step of Sry action has been discovered, where Sry together with Sf-1 binds to a testis-specific enhancer element (TESCO). Sox9 upregulation during testis development is highly conserved and observed in both mammals and birds (Smith et al. 1999). Sp1 binding sites have been identified for Sox9 (Piera-Velazquez et al. 2007; Bagheri-Fam et al. 2010), raising a potential role for Crspx/y to modulate expression of Sox9. Using the mouse enhancer element and a range of genes including Sox3, we did not observe a significant effect on the reporter gene. Modest activation of the reporter by both platypus genes do demonstrate that platypus Crspx and Crspy have an effect in this system. Importantly, this shows that it is feasible to test platypus genes using this approach, and in future work we will use increasingly platypus-specific sequences including the platypus Sox9 enhancer element and platypus orthologs of Sox3, Sox9, and Sf-1. This may reveal a possible role of Crspx and Crspy in testis, but also help understanding the evolutionary dynamic of Sox9 regulation in mammalian evolution before the origin of the Sry gene.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- CDYL :
-
Chromodomain protein, Y chromosome-like, processed pseudogene
- CDY :
-
Chromodomain protein, Y-linked
- DMRT1 :
-
Doublesex and mab-3 related transcription factor 1
- DMY :
-
Y-specific DM-domain gene required for male development in the medaka fish
- MED15:
-
Mediator complex subunit 15
- CRSP7 :
-
(MED26) Mediator of RNA polymerase II transcription subunit 26. Cofactor required for Sp1 transcriptional activation, subunit 7
- Crspx :
-
Mediator complex subunit 26 on platypus X
- Crspy :
-
Mediator complex subunit 26 on platypus Y
- SF-1 :
-
Steroidogenic factor 1
- SOX9 :
-
SRY (sex-determining region Y)-box 9
- SP1 :
-
Specific protein 1 (transcription factor)
- SRY :
-
Sex-determining region on Y
- TESCO :
-
Testis-specific enhancer core element
- UPGMA:
-
Unweighted Pair Group Method with Arithmetic Mean
References
Ashley T (2002) X-autosome translocations, meiotic synapsis, chromosome evolution and speciation. Cytogenet Genome Res 96:33–39
Ashley T (2005) Chromosome chains and platypus sex: kinky connections. Bioessays 27:681–684
Bagheri-Fam S, Sinclair AH, Koopman P, Harley VR (2010) Conserved regulatory modules in the sox9 testis-specific enhancer predict roles for sox, tcf/lef, forkhead, dmrt, and gata proteins in vertebrate sex determination. Int J Biochem Cell Biol 42:472–477
Berlin S, Ellegren H (2006) Fast accumulation of nonsynonymous mutations on the female-specific w chromosome in birds. J Mol Evol 62:66–72
Bick YA, Murtagh C, Sharman GB (1973) The chromosomes of an egg-laying mammal tachyglossus aculeatus (the echidna). Cytobios 7:233–243
Charlesworth B (1991) The evolution of sex chromosomes. Science 251:1030–1033
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118–128
Cioffi MB, Molina WF, Moreira-Filho O, Bertollo LA (2011a) Chromosomal distribution of repetitive DNA sequences highlights the independent differentiation of multiple sex chromosomes in two closely related fish species. Cytogenet Genome Res 134:295–302
Cioffi MB, Sanchez A, Marchal JA, Kosyakova N, Liehr T, Trifonov V et al (2011b) Cross-species chromosome painting tracks the independent origin of multiple sex chromosomes in two cofamiliar erythrinidae fishes. BMC Evol Biol 11:186
Desclozeaux M, Poulat F, de Santa Barbara P, Soullier S, Jay P, Berta P et al (1998) Characterization of two sp1 binding sites of the human sex determining sry promoter. Biochim Biophys Acta 1397:247–252
Djakiew D, Jones RC (1982) Ultrastructure of the ductus epididymidis of the echidna, tachyglossus aculeatus. J Anat 135:625–634
Djakiew D, Jones RC (1983) Sperm maturation, fluid transport, and secretion and absorption of protein in the epididymis of the echidna, tachyglossus aculeatus. J Reprod Fertil 68:445–456
Dohm JC, Tsend-Ayush E, Reinhardt R, Grutzner F, Himmelbauer H (2007) Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes. Genome Biol 8:R175
Dorus S, Gilbert SL, Forster ML, Barndt RJ, Lahn BT (2003) The cdy-related gene family: coordinated evolution in copy number, expression profile and protein sequence. Hum Mol Genet 12:1643–1650
Edgar RC (2004) Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ellegren H (2011) Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet 12:157–166
Felsenstein J (1974) The evolutionary advantage of recombination. Genetics 78:737–756
Galian J, Proenca SJ, Vogler AP (2007) Evolutionary dynamics of autosomal-heterosomal rearrangements in a multiple-x chromosome system of tiger beetles (cicindelidae). BMC Evol Biol 7:158
Gruetzner F, Ashley T, Rowell DM, Marshall Graves JA (2006) How did the platypus get its sex chromosome chain? A comparison of meiotic multiples and sex chromosomes in plants and animals. Chromosoma 115:75–88
Grutzner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O’Brien PC, Jones RC et al (2004) In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird z and mammal x chromosomes. Nature 432:913–917
Grutzner F, Nixon B, Jones RC (2008) Reproductive biology in egg-laying mammals. Sex Dev 2:115–127
Huelsenbeck JP, Ronquist F (2001) Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ et al (2006) Chimpanzee and human y chromosomes are remarkably divergent in structure and gene content. Nature 463:536–539
Jones RC, Djakiew D, Dacheux JL (2004) Adaptations of the echidna (Tachyglossus aculeatus) for sperm production, particularly in an arid environment. Australian Mammalogy 26:199–204
Jones RC, Dacheux JL, Nixon B, Ecroyd HW (2007) Role of the epididymis in sperm competition. Asian J Androl 9:493–499
Kaiser VB, Bachtrog D (2010) Evolution of sex chromosomes in insects. Annu Rev Genet 44:91–112
Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC et al (2007) Pycogent: a toolkit for making sense from sequence. Genome Biol 8:R171
Kortschak RD, Tsend-Ayush E, Grutzner F (2009) Analysis of sine and line repeat content of y chromosomes in the platypus, ornithorhynchus anatinus. Reprod Fertil Dev 21:964–975
Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25
Malik S, Roeder RG (2010) The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11:761–772
Marshall Graves JA (2008) Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet 42:565–586
Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, Paul-Prasanth B et al (2007) Dmy gene induces male development in genetically female (xx) medaka fish. Proc Natl Acad Sci USA 104:3865–3870
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Annu Rev Plant Biol 62:485–514
Muller HJ (1918) Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3:422–499
Murphy WJ, Pearks Wilkerson AJ, Raudsepp T, Agarwala R, Schaffer AA, Stanyon R et al (2006) Novel gene acquisition on carnivore y chromosomes. PLoS Genet 2:e43
Murtagh C (1977) A unique cytogenetics system in monotremes. Chromosoma 65:37–57
Niculita-Hirzel H, Stock M, Perrin N (2008) A key transcription cofactor on the nascent sex chromosomes of european tree frogs (Hyla arborea). Genetics 179:1721–1723
Piera-Velazquez S, Hawkins DF, Whitecavage MK, Colter DC, Stokes DG, Jimenez SA (2007) Regulation of the human sox9 promoter by sp1 and creb. Exp Cell Res 313:1069–1079
Rens W, Grutzner F, O’Brien PC, Fairclough H, Graves JA, Ferguson-Smith MA (2004) Resolution and evolution of the duck-billed platypus karyotype with an x1y1x2y2x3y3x4y4x5y5 male sex chromosome constitution. Proc Natl Acad Sci USA 101:16257–16261
Rens W, O’Brien PC, Grutzner F, Clarke O, Graphodatskaya D, Tsend-Ayush E et al (2007) The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian z. Genome Biol 8:R243
Rice WR (1987) Genetic hitchhiking and the evolution of reduced genetic activity of the y sex chromosome. Genetics 116:161–167
Rowell DM (1987) Complex sex-linked translocation heterozygosity: its genetics and biological significance. Trends Ecol Evol 2:242–246
Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH et al (2003) Abundant gene conversion between arms of palindromes in human and ape y chromosomes. Nature 423:873–876
Ryu S, Zhou S, Ladurner AG, Tjian R (1999) The transcriptional cofactor complex crsp is required for activity of the enhancer-binding protein sp1. Nature 397:446–450
Sandstedt SA, Tucker PK (2005) Male-driven evolution in closely related species of the mouse genus mus. J Mol Evol 61:138–144
Sekido R, Lovell-Badge R (2008) Sex determination involves synergistic action of sry and sf1 on a specific sox9 enhancer. Nature 453:930–934
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG et al (2003) The male-specific region of the human y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837
Smith CA, Smith MJ, Sinclair AH (1999) Gene expression during gonadogenesis in the chicken embryo. Gene 234:395–402
Stock M, Horn A, Grossen C, Lindtke D, Sermier R, Betto-Colliard C et al (2011) Ever-young sex chromosomes in european tree frogs. PLoS Biol 9:e1001062
Tsend-Ayush E, Dodge N, Mohr J, Casey A, Himmelbauer H, Kremitzki CL, Schatzkamer K, Graves T, Warren WC, Grützner F. (2009) Higher-order genome organization in platypus and chicken sperm and repositioning of sex chromosomes during mammalian evolution. Chromosoma. 118(1):53-69
Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE et al (2011) Human mediator subunit med26 functions as a docking site for transcription elongation factors. Cell 146:92–104
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE et al (2008) Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18:965–973
Wallis MC, Waters PD, Delbridge ML, Kirby PJ, Pask AJ, Grutzner F et al (2007) Sex determination in platypus and echidna: autosomal location of sox3 confirms the absence of sry from monotremes. Chromosome Res 15:949–959
Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F et al (2008) Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183
Wilson MA, Makova KD (2009) Evolution and survival on eutherian sex chromosomes. PLoS Genet 5:e1000568
Wyckoff GJ, Li J, Wu CI (2002) Molecular evolution of functional genes on the mammalian y chromosome. Mol Biol Evol 19:1633–1636
Acknowledgments
We thank Dr. Paul Thomas for sharing unpublished results for Sox3 and for sharing the Sox3 expression construct. E.T-A and F.G. acknowledge funding from the Australian Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: Tariq Ezaz and Jennifer Graves.
Enkhjargal Tsend-Ayush and R. Daniel Kortschak contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Crsp7 multiple alignment used in the phylogenetic analysis (Fig. 1). Species included in the analysis were zebrafish (D. rerio), frog (X. laevis), chicken (G. gallus), opossum (M. domestica), human (H. sapiens), and platypus (O. anatinus), Crspx and Crspy (JPEG 344 kb)
Supplementary Fig. 2
Evolution of the sex chromosome chain from a ring. This alternative model is discussed in Gruetzner et al. (2006) in detail. The homology between X1 and Y5 reported here is the first evidence that a ring structure may have existed in monotremes (JPEG 30 kb)
ESM 1
Supplementary data: FASTA sequences of Crsp7 sequences used in the phylogenetic analysis (FASTA 14 kb)
Rights and permissions
About this article
Cite this article
Tsend-Ayush, E., Kortschak, R.D., Bernard, P. et al. Identification of mediator complex 26 (Crsp7) gametologs on platypus X1 and Y5 sex chromosomes: a candidate testis-determining gene in monotremes?. Chromosome Res 20, 127–138 (2012). https://doi.org/10.1007/s10577-011-9270-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9270-z