Abstract
The accurate segregation of duplicated chromosomes, essential for the development and viability of a eukaryotic organism, requires the formation of a robust microtubule (MT)-based spindle apparatus. Entry into mitosis or meiosis precipitates a cascade of signalling events which result in the activation of pathways responsible for a dramatic reorganisation of the MT cytoskeleton: through changes in the properties of MT-associated proteins, local concentrations of free tubulin dimer and through enhanced MT nucleation. The latter is generally thought to be driven by localisation and activation of γ-tubulin-containing complexes (γ-TuSC and γ-TuRC) at specific subcellular locations. For example, upon entering mitosis, animal cells concentrate γ-tubulin at centrosomes to tenfold the normal level during interphase, resulting in an aster-driven search and capture of chromosomes and bipolar mitotic spindle formation. Thus, in these cells, centrosomes have traditionally been perceived as the primary microtubule organising centre during spindle formation. However, studies in meiotic cells, plants and cell-free extracts have revealed the existence of complementary mechanisms of spindle formation, mitotic chromatin, kinetochores and nucleation from existing MTs or the cytoplasm can all contribute to a bipolar spindle apparatus. Here, we outline the individual known mechanisms responsible for spindle formation and formulate ideas regarding the relationship between them in assembling a functional spindle apparatus.
Similar content being viewed by others
Part I: The mechanisms underpinning spindle assembly
Centrosome-nucleated MTs
Early experiments in animal cells unequivocally established centrosomes as the primary microtubule organising centres (MTOCs) during mitosis (see Varmark 2004 for review). For example, unfertilised Xenopus embryos, which do not ordinarily contain a centrosome, will undergo cell cycle oscillations when pricked with a needle. However, these embryos will not undergo cell division unless a centrosome is co-injected (Maller et al. 1976). Centrosomes consist of a pair of perpendicular centrioles that organise the surrounding pericentriolar material (PCM; Bobinnec et al. 1998). The PCM is an electron-dense structure consisting of many proteins, including γ-tubulin, probably in the form of the γ-tubulin ring complex (γ-TuRC; Hannak et al. 2002). Increased nucleation of microtubules (MTs) by centrosomes at the onset of mitosis or meiosis depends on the coordinated action of centriolar and PCM components that recruit and anchor increased amounts of γ-tubulin at the spindle pole. In human cells, this is facilitated, at least in part, by the centrosomal protein, Pericentrin (Zimmerman et al. 2004). In flies, where the pathway of centrosome maturation has been more extensively studied, the Pericentrin-like protein (D-PLP) shares this role with centrosomin (Cnn; Martinez-Campos et al. 2004). The localisation of both these PCM proteins is dependent upon the presence of the core centriolar component D-Spd2 (Dix and Raff 2007; Giansanti et al. 2008), whilst the phosphorylation of Cnn by the mitotic kinase, Polo, correlates with centrosome maturation (Dobbelaere et al. 2008), suggesting a hierarchical pathway in which γ-tubulin is targeted and maintained at the PCM in a stepwise fashion.
Interestingly, centrosome maturation does not fully account for the increase in astral MTs nucleated early in mitosis. Centrosomal MTs, once formed, do not remain tightly bound, but are nonetheless held in the vicinity of the PCM (Mastronarde et al. 1993; Wakefield et al. 2000; Varmark 2004). Whether the detachment of newly nucleated MTs from the γ-TuRC template is a passive process or whether an active severing of MTs close to the minus end occurs, it is clear that maintaining an interaction between MTs and the PCM is crucial for centrosome function; the loss of microtubule-associated protein (MAPs) such as NuMA, in mammalian cells, and Asp, in flies, leads to centrosomes that continue to nucleate short astral MTs, but that detach from the bulk of MTs that have already contributed to the growing spindle (Gonzalez et al. 1990; Merdes et al. 1996; Wakefield et al. 2001).
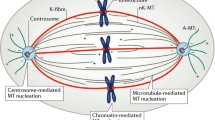
The dramatic increase in centrosome-driven MT nucleation at the onset of mitosis described above leads to the astral arrays so familiar in textbooks (Fig. 1). Although initially radial, the centrosomally derived MTs begin to grow with directional bias such that the density of MTs between the centrosomes and the mitotic chromatin is greater than between the centrosomes and the cell cortex. It is currently unclear which mechanisms precipitate this directionality though a gradient of MT-stabilising activity affecting MAPs and MT motor proteins at mitotic chromosomes is likely to be important.
Despite the well-characterised function of centrosomes during spindle formation, it is clear that higher plants do not employ this mechanism (see article from Zhang and Dawe in this issue; Shimamura et al. 2004; Brown and Lemmon 1997). In addition, there is much accumulated evidence of acentrosomal meiotic systems in animal cells, such as mouse (Calarco-Gillam et al. 1983), Xenopus (Heald et al. 1996) and Drosophila (Matthies et al. 1996). Furthermore, physical displacement of centrosomes from the nucleus in crane fly spermatocytes, or laser ablation of both centrosomes during prophase in monkey CVG-2 cells by laser microsurgery, does not disrupt bipolar spindle assembly (Steffen et al. 1986; Khodjakov et al. 2000). The observation that cells within a developing animal which normally contain centrosomes could, in fact, build a functional spindle in the complete absence of these organelles was confirmed genetically through removing, first core centrosomal and then core centriolar, proteins in fruit flies. Drosophila cnn-null mutants fail to recruit PCM, thus rendering centrosomes functionally inactive. Nonetheless, spindles in cnn-null mutants are formed in the absence of astral MTs, and these flies develop to adulthood with few observable defects in morphology or developmental timing (Megraw et al. 2001). Moreover, a null mutant in DSas-4, a centriole duplication factor, develops into a morphologically normal adult. Although severe coordination defects due to a lack of cilia are ultimately lethal for these adults and larval brain cells commonly show both a delay in mitosis and a failure to establishing asymmetry, bipolar mitotic spindles capable of chromosome segregation form (Basto et al. 2006). Thus, it is clear that although when present centrosomes dominate, they are not essential for spindle assembly.
Ran-dependent, chromatin-directed MT generation
The ability of cells to build a spindle in the absence of centrosomes implies that redundant mechanisms must exist to generate and organise MTs. An alternative route to spindle formation in animal cells has been described most clearly in an in vitro system using Xenopus egg extracts. When sperm nuclei (containing centrosomes) are added to such extracts, focused spindles form primarily via centrosomal nucleation of MTs followed by the concerted action of motor proteins such as cytoplasmic dynein and MAPs, such as NuMA (Sawin and Mitchison 1991; Heald et al. 1997). However, the addition of beads coated with mitotic chromatin to activated extracts lacking centrosomes also initiates the formation of spindles with two focussed poles possessing a similar distribution of spindle lengths as those with centrosomes (Heald et al. 1996) (Fig. 1). Interestingly, however, these spindles take much longer to form and pass through a number of intermediate structures not seen when centrosomes are present.
This chromatin-directed MT assembly is dependent upon the small GTPase Ran (Carazo-Salas et al. 1999; Kalab et al. 1999) via the localisation of its guanine nucleotide exchange factor, RCC1 (Moore et al. 2002; Li et al. 2003). Upon nuclear envelope breakdown, RCC1 binds to condensing chromatin. This generates a well-characterised gradient of Ran.GTP in the vicinity of the chromosomes. First described in the Xenopus in vitro system, using FRET reporters (Kalab et al. 1999), this gradient has also been shown to be present in human somatic cells (Kaláb et al. 2006). MT organisation by Ran.GTP relies upon its liberation of cargo proteins from the nuclear import factor Importin-β (reviewed by Ciciarello et al. 2007). These liberated cargoes, termed spindle assembly factors, include TPX2, a spindle pole protein required for the targeting of XKlp2 to MT minus ends (Wittmann et al. 2000; Gruss et al. 2001) and the MT-associated protein HURP (Wong et al. 2006). TPX2 induces aster formation at the spindle poles via a physical interaction with Aurora A, contributing to the activation of the kinase (Trieselmann et al. 2003). Active Aurora A then stabilises MTs via a protein complex containing Msps/XMAP215, Eg5 and HURP (Koffa et al. 2006).
However, in the absence of pre-polymerised MTs, HURP has also been shown to act to stabilise MTs independently of this complex in a Ran.GTP-dependent manner (Casanova et al. 2008). Moreover, Xenopus extracts with the Ran.GTP gradient flattened can support bipolar spindle formation when sperm nuclei (containing kinetochores and centrosomes) are added, thus implicating additional spatial signals for guiding MT growth (Maresca et al. 2009a).
Kinetochore-driven MT generation
For over 35 years, it has been known that kinetochores can initiate MT polymerisation independently of centrosomes in mammalian tissue culture cells (McGill and Brinkley 1975; Snyder and McIntosh 1975). Live imaging of MTs has conclusively shown that in normal mitotic cells, these kinetochore-generated MTs exist, incorporating into the growing, centrosome-driven spindle in a dynein-dependent process via their capture by astral MTs (Khodjakov et al. 2003; Maiato et al. 2004; see Rieder 2005 for review) (Fig. 1).
However, the mechanisms underlying kMT generation, and their contribution to spindle assembly in normal cells, have been difficult to elucidate. This is mainly due to the relationship between MTs and kinetochores. In the classical search–capture model of chromosome alignment (Kirchner and Mitchison 1986; Nicklas et al. 1994), astral MTs from opposing centrosomes attach to kinetochores, facilitating chromosome congression. But if MTs from the same pole attach to sister kinetochores, the interaction is selectively destabilised such that “correct” MT–kinetochore relationships can be established. Thus, kinetochores must contain MT binding sites and act as a readout of merotely and syntely, in addition to themselves generating MTs.
The mechanism by which kinetochores themselves nucleate MTs appears to be, like general chromatin-mediated MT generation, dependent on Ran.GTP. Injection of Importin-β and depletion of either TPX2 or HURP both diminish MT formation at kinetochores following MT depolymerisation (Tulu et al. 2006; Yang and Fan 2008), whilst the fraction of Ran.GTP present at kinetochores appears to be responsible for nucleating kinetochore-associated MTs (Torosantucci et al. 2008). However, live analyses of cells undergoing mitosis with unreplicated genomes (MUG) complicate this interpretation. Following treatment with hydroxyurea, kinetochores become uncoupled from chromosomes, forming a bipolar spindle, whilst unreplicated chromatin is excluded from the spindle (Brinkley et al. 1988; O’Connell et al. 2008). During MUG, RCC1 binds chromatin, but not kinetochores, suggesting that kinetochores can support bipolar spindle formation without the direct influence a Ran.GTP signal (O’Connell et al. 2009). Thus, a precise relationship between Ran.GTP and kinetochore-driven MT generation remains ambiguous.
CPC-directed MT generation
The chromosomal passenger complex (CPC)—consisting of Aurora B, INCENP, Survivin and Borealin/Dasra-B—is required for spindle bipolarity, chromosome alignment and cytokinesis. Its presence at the chromosome arms during early prometaphase implicates it in MT stabilisation; its re-localisation to the inner centromeres at late prometaphase enables it to regulate the stability of kinetochore–kMT interactions (for review, see Ruchaud et al. 2007), whilst its presence along the spindle suggests a more general role in MT generation during spindle formation (Tseng et al. 2010; our unpublished observations).
Undoubtedly, the CPC has a role in modulating MT dynamics to facilitate spindle bipolarity. Op18/Stathmin inhibits MT polymerisation in vitro and is negatively regulated in the vicinity of chromatin during spindle formation (Andersen et al. 1997). Inhibition of Op18/Stathmin depends on hyperphosphorylation by Aurora B (Gadea and Ruderman 2006), whilst chromatin beads cannot form bipolar spindles in the absence of CPC activity in Xenopus extracts—an effect that is reversed by co-depletion of INCENP with the MT depolymerising enzyme MCAK (Sampath et al. 2004). However, there is increasing evidence that the CPC has an additional direct role in generating a population of MTs that incorporate into the growing spindle. Reconstitution of active CPC on beads supports MT assembly in the absence of Ran.GTP (Kelly et al. 2007), whilst sperm nuclei which generate bipolar spindles in the absence of a Ran.GTP gradient cannot, however, promote local MT generation after either INCENP immunodepletion or chemical inhibition of Aurora B in this system (Maresca et al. 2009b). Furthermore, the association of active Aurora B with spindle MTs (as opposed to kinetochores) is essential for spindle assembly, with spindle-associated Aurora B effectively transmitting a spatial signal from chromosomes to promote bipolar spindle formation (Tseng et al. 2010).
Whatever the precise mechanisms, the importance of MT organisation by the CPC in Drosophila is demonstrated by a drastic delay in female meiotic spindle formation in incenp mutants (Colombié et al. 2008), suggesting a conserved role for the CPC in directing MT generation during spindle formation.
Augmin-generated MTs
Both fluorescence-speckled microscopy of anastral meiotic spindles in Xenopus and live cell imaging of the plus end-associated protein, EB1, in cells without functional centrosomes have revealed a distribution of MT ends throughout the spindle (Burbank et al. 2006; Mahoney et al. 2006), suggesting that new MTs are generated at sites distant from centrosomes or chromatin during mitosis.
Indeed, recent work has uncovered a set of MAPs, originally identified in Drosophila, that appear to have a role in MT generation from within the spindle (Brown and Lemmon 1997, 2008; Hughes et al. 2008). These proteins localise a subpopulation of γ-tubulin to existing spindle MTs, and their absence leads to spindles of lower density with an increased astral population (Goshima et al. 2008; Hughes et al. 2008). Termed Augmin, or HAUS (homologous to Augmin subunits; Lawo et al. 2009; Uehara et al. 2009), this conserved hetero-octomeric complex has been shown to directly interact with the γTuRC subunit, NEDD1 (Zhu et al. 2008), a result that has been independently verified (Teixidó-Travesa et al. 2010). Thus, a model has been proposed in which Augmin binds existing spindle MTs during prometaphase and targets active γTuRC, facilitating intraspindle MT nucleation (Goshima et al. 2008; Uehara et al. 2009) (Fig. 1).
Similarly to the centrosomal contribution of MTs to spindle formation, MTs generated by Augmin are dispensable for animal development. Null mutants of the Augmin subunit, Wac, are viable, albeit showing a small, but significant, impact on somatic mitosis (Meireles et al. 2009). However, Augmin is essential for the fast and synchronous early embryonic divisions that occur in the embryo of the fly. Mitotic spindles from embryos laid by mothers mutant for the gene encoding the Augmin subunit, msd1, are larger and less dense than their wild-type counterparts, failing in chromosome alignment and segregation (Wainman et al. 2009).
Although Augmin has currently only been characterised in Drosophila and humans, it is likely that it exists in other species. Its identification is complicated by the poor conservation at the amino acid level; only regions of four of the eight subunits show significant homology between humans and Drosophila, with some subunits appearing to have homologues in Xenopus and Zebrafish (Uehara et al. 2009). However, de novo homology modelling of the other subunits suggests conservation at the level of tertiary structure; three of the four Augmin proteins from either species appear to structurally pair with each other (Fig. 2). Furthermore, the demonstration that MT minus ends are present throughout the Xenopus spindle (Burbank et al. 2006), together with recent mathematical modelling of Xenopus meiotic spindle formation in which spindle-driven MT amplification was incorporated (Loughlin et al. 2010), suggests that this mechanism plays a crucial role in spindle formation.
Mechanisms known to contribute to mitotic spindle formation. Whilst the centrosome-nucleated pathway dominates in cells containing these organelles, kinetochore- and chromatin-driven MT generation additionally occurs, the latter mediated by both Ran.GTP- and CPC-mediated mechanisms. Current evidence suggests that a pathway involving the Augmin complex has a role in generating MTs from preexisting spindle MTs, increasing its density, whilst in some cells, MT formation from the region of the nuclear envelope and from de novo cytoplasmic foci of γ-tubulin has been reported. Together, these mechanisms combine to ensure robust spindle formation even in the absence of one or more pathways
Other mechanisms of MT generation
Theoretically, any cellular structure that sequesters active MT nucleators could act as a focus for MT generation during spindle formation. Whilst centrosomes, chromatin and existing MTs act as the dominant anchors, there is evidence for additional sites of MT nucleation (Fig. 1). For example, the remnants of the nuclear envelope and the proposed spindle matrix are both candidates, being partly composed of Nuceloporin (Nup) proteins, some of which have been shown to associate with γ-tubulin (see review by Johansen et al., in this issue). In addition, work in Drosophila spermatocytes in which centrosomes have been genetically displaced from their normal juxtanuclear position clearly shows MT amplification occurring in the region of the nuclear envelope (Rebollo et al. 2004), whilst a recent investigation into spindle formation in mouse oocytes also reported the presence of perinuclear MTs contributing to spindle assembly (Schuh and Ellenberg 2007). Further evidence for nucleation of MTs from membranous structures comes from land-based plants where the generation of a cortical pre-prophase band of MTs occurs upon entry into mitosis (Chan et al. 2005; see review by Zhang and Dawe in this issue) and where γ-tubulin associates directly with the nuclear envelope in mitosis (for example, Smirnova and Bajer 1994) and with membranous plastids during meiosis (Brown and Lemmon 1997; Shimamura et al. 2004).
The cytoplasm, too, is able to generate MTs that contribute to spindle formation, seemingly independently of any symmetry-breaking structure. Haemanthus endosperm cytoplasts (plant cell fragments devoid of nuclei and centrosomes) form large regular arrays of MTs upon entry into mitosis which can arrange into chromosome free mitotic spindles (Bajer and Mole-Bajer 1986), whilst mouse oocyte cytoplasts nucleate asters of MTs which progressively interact with one another, again forming bipolar spindles (Brunet et al. 1998). Recent work has shown that over 80 such MTOCs exist in intact mouse oocytes undergoing cell division (Schuh and Ellenberg 2007), whilst similar acentrosomal MTOCs (aMTOCs), containing both γ-tubulin and Cnn, have also been observed in Drosophila cells devoid of centrosomes, where they contribute substantially to spindle assembly (Mottier-Pavie and Megraw 2009). Together, these studies suggest that cytoplasmic MT generation during mitosis and meiosis is a widespread phenomenon (Fig. 2).
The predicted tertiary structures of Drosophila and human Augmin subunits are similar despite the absence of significant amino acid homology. Both Drosophila and human Augmin have been shown to contain eight subunits. Of these, four are conserved between species at the amino acid level. The remaining eight amino acid sequences (Drosophila Msd1, Wac, Dgt2 and Dgt8, and human Cep27, Ccdc5, UCHL5IP1 and C14orf94) were subjected to de novo tertiary structure prediction software (I-TASSER; Zhang 2008). Pairwise combinations of the best-fit structures were compared using DALI (Holm et al. 2008), where scores over 2.0 are considered significant. No significant predicted homology was found between the best-fit structures of Dgt2 and UCHL5IP1. However, three pairs of Drosophila–human structures were identified with high confidence, suggesting that Msd1, Wac and Dgt8 are the orthologues of Cep27, Ccdc5 and C14orf94, respectively
Part II: Relative contributions of pathways to spindle formation
It is clear that certain mechanisms of MT generation during spindle assembly dominate in certain cells. However, it does not necessarily follow that particular cells only use a subset of these mechanisms. As described earlier, removal of centrosomes from cells genetically, through laser ablation or through RNAi, reveals the concomitant presence of chromatin-dependent MT nucleation (Khodjakov et al. 2000; Mahoney et al. 2006). Moreover, the removal of the dominant Ran-driven MT generation pathway in Xenopus extracts uncovered the CPC as an additional regulator of MT generation (Kelly et al. 2007). The ability of a single cell to use multiple pathways during spindle formation is further supported by genetic studies in which two mechanisms have been simultaneously disrupted; flies lacking either the core centrosomal protein Cnn (and therefore centrosome-driven MT generation) or the Augmin subunit Msd1 survive to adulthood and form spindles capable of satisfying the SAC. When both pathways are removed, spindle formation is severely compromised, leading to organismal lethality (Wainman et al. 2009), supporting a model in which both pathways contribute distinct populations of MTs. However, MTs are still present in these highly disrupted mitoses, accumulating around the mitotic chromatin (Wainman et al. 2009). Thus, it seems likely that multiple mechanisms exist in most cells, but that some contribute more to spindle formation than others. So the question is, what determines the preference of the cell to use one pathway to a greater extent than the others?
Spatial regulation of MT nucleation
One possibility is that pathway preference is determined simply by the number of available MT-generating sites within the growing spindle. For example, the accumulation of γ-TuRC at specific locations will result in the nucleation of MTs from these sites. Thus, the dramatic increase in PCM and associated γ-tubulin at the centrosome seen upon entry into mitosis in most animal cells will directly correlate with the increased nucleating capacity of the centrosome. Similarly, the increase of Augmin binding sites as spindle MTs are newly generated during prometaphase could lead to a proportional increase in γ-tubulin to these MTs and a subsequent increase in MT generation via this pathway. Extending this hypothesis to chromatin or nuclear envelope-mediated MT generation, one could argue that the more chromosomal DNA or kinetochores a cell possesses, or the larger the nucleus, the more available nucleation sites would be available and the more these pathways would dominate over others.
Such a direct mechanism could be capable of greater flexibility if the availability of nucleating sites were regulated. This could be achieved either by holding a single MT nucleator (i.e., γ-tubulin) inactive at certain subcellular locations, such that one mechanism predominates, or through regulating a number of different MT nucleators independently of each other. The presence of γ-tubulin complexes in the cytoplasm, which are able to incorporate into foci of MT-generating aMTOCs (Mottier-Pavie and Megraw 2009), is strong evidence for the regulation of γ-tubulin-dependent nucleation. However, the absence of γ-tubulin on mitotic chromatin (except at the kinetochore in certain circumstances, see below) suggests that other proteins may be capable of MT nucleation and therefore may play a role in generating MTs during spindle assembly. Indeed, TPX2 alone appears to nucleate MTs, promote aster formation and bundle MTs in pure tubulin solutions (Schatz et al. 2003), a process which is enhanced by the addition of the TPX2 binding partner XRHAMM (Groen et al. 2004). Another Ran.GTP-dependent MAP, ISWI, also has MT nucleation and bundling activity in pure tubulin solutions (Yokoyama et al. 2009), whilst work in interphase Drosophila tissue culture cells has led to the suggestion that MTs may be nucleated in the absence of γ-tubulin through interaction between tubulin heterodimers and a complex of MAPs, including EB1, Msps and CLIP-190 (Rogers et al. 2008). Furthermore, MTs need not be nucleated in order to increase in number during mitosis; MT severing provides an alternative potential mechanism of increasing the number of MTs in the absence of γ-tubulin-driven nucleation. Indeed, severing enzymes, such as Katanin in Caenorhabditis elegans, and its orthologue, KL1 in humans, have been proposed to generate new MT seeds during spindle formation (Sonbuchner et al. 2010; Srayko et al. 2006).
Temporal regulation of MT generation
As with any dynamic cellular process, temporal restrictions are as important as spatial constraints. It is likely that the variation in pathway usage seen in spindle formation is at least partially due to a partitioning of tubulin, MT nucleators and their binding sites, a partitioning that alters over time. The activation of mitotic cdk-cyclin upon entry into mitosis leads to a cascade of increased MT nucleation. However, in many cells, this initial increase in cytoplasmic activity occurs prior to onset of nuclear envelope breakdown (NEB), and thus the pool of αβ-tubulin (and indeed γ-tubulin) is itself primarily confined to the cytoplasm. Consequently, any MT nucleating sites within the nucleus—for example on chromatin or kinetochores—will not be able to generate MTs. In contrast, centrosomes and the nuclear envelope itself sit outside of this physical barrier, surrounded by tubulin from the very onset of mitosis. This could explain why, when present, astral MTs generated by the centrosome initially predominate as the main source of spindle MTs, perhaps masking concomitant nuclear envelope- and cytoplasmic-generated MTs. Subsequent to NEB, however, chromatin- and kinetochore-driven MT generation would be initiated, leading to a second burst of MT generation. In such a model, Augmin-dependent MT generation would be biphasic, initially acting to form new MTs from centrosomally derived spindle MTs, before using the MTs synthesised by chromatin.
Certainly, this is consistent with what is seen in land-based plants lacking centrosomes and Drosophila spermatocytes in which centrosomes have been displaced, where MT generation around the nuclear envelope is visible prior to NEB (see above). One further observation that supports such a temporally restricted model is based on the dynamics of spindle assembly in centrosome-containing cells after transient MT depolymerisation, brought about either by incubation with MT inhibitors or through being placed at 4°C. In both cases, cells progress past NEB, but cannot satisfy the spindle assembly checkpoint, arresting with condensed chromatin in a prometaphase-like state. Upon drug removal or rewarming, respectively, such cells will produce normal mitotic spindles capable of chromosome segregation (see Inoue and Sato 1967). However, the dynamics of spindle assembly differ from untreated cells. Instead of building their spindles via MT generation from the centrosomes, spindle formation proceeds primarily from kinetochores and/or chromatin, with the centrosomes initially contributing only a minor proportion (see, for example, Torosantucci et al. 2008; Bucciarelli et al. 2009). This correlates with an enrichment of kinetochore-associated γ-tubulin, which is normally only very weakly present at kinetochores during spindle formation, and, additionally in Drosophila tissue culture, Augmin, which is normally absent (Torosantucci et al. 2008; Bucciarelli et al. 2009). These results can be adequately explained by the temporal activation model described above; cold-treated mitotic cells progress through the normal cascades associated with cdk-cyclin activation such that they end up in a “metaphase-like” cytoplasmic state, with no nuclear–cytoplasmic boundary, and with “primed” MT nucleating sites at the kinetochores, but they cannot begin MT generation. Once the temperature restriction has been removed, however, spindle formation occurs via a “free for all”, with each mechanism competing for the available tubulin.
The temporal model of spindle formation may also suffice, at least partly, as an explanation of the different extents to which Augmin-generated MTs contribute to spindle formation in different Drosophila tissues. Genetic removal of Augmin from larval neuroblasts appears to have no effect on the shape or size of the mitotic spindle, as assessed by indirect immunofluorescence of metaphase cells (Meireles et al. 2009; Wainman et al. 2009). In contrast, the mitotic spindles of syncytial blastoderm embryos laid by this same mutant are less dense and longer than their wild-type counterparts, with apparently larger MT asters (Wainman et al. 2009). However, a key difference in spindle formation between these two cell types is the strength of the spindle assembly checkpoint. In neuroblasts, the checkpoint ensures that anaphase proceeds only after the formation of a robust spindle, with a full complement of kinetochore–kMT attachments. In contrast, the syncytial nature of the early embryo can result in a “communal” progression through mitosis where the status of the majority of spindles overrides individual nuclei. In msd1 mutant embryos, although the initial burst of MT generation from the centrosomes occurs as in wild type, NEB and robust spindle formation are delayed in some nuclei. Interestingly, during this delay, MT generation also appears to occur around the corresponding nuclear envelopes (our unpublished observations). However, these spindles continue through, and exit, mitosis with only a minor delay, manifesting the comparatively weak spindle density phenotype. In contrast, the delay in spindle formation due to loss of Augmin in neuroblasts will be compensated for by the increased time spent in prometaphase, which will allow the other MT-generating pathways (e.g. chromatin-mediated) to gradually contribute to spindle formation.
Integrated regulation of spindle formation pathways
The above observations suggest that spindle formation occurs in a partitioned and stepwise manner, with the sequential activation of a number of distinct pathways of MT generation, dependent upon the local availability of required resources.
However, one intriguing possibility, which could also contribute to the differential activity of individual mechanisms seen in different cells, is that the pathways of MT generation positively or negatively regulate each other. Certainly, some intrinsic regulation exists; spindle formation does not persist unabated throughout mitosis. Instead, a steady state is reached in which a mature mitotic spindle is still in an environment supporting MT generation, but does not continue to nucleate MTs to the same extent (this is perhaps best visualised in a thought experiment for Augmin which uses preexisting spindle MTs to generate new ones. Without regulation, this would be expected to generate and maintain an exponential increase in spindle MTs over time—something that clearly does not happen). Thus, feedback mechanisms regulating the overall density of the mitotic spindle must exist.
Indeed, there is evidence of cross talk between pathways; for example, when centrosomes are removed from a cell normally containing them, the dynamics of kinetochore- and chromatin-driven generation increases (Torosantucci et al. 2008; our unpublished observations). Although this could be attributed to increased resources being available for a subsequently activated pathway upon removal of a preceding one (as described in the previous section), this would not account for other observations; when Augmin-dependent MTs are reduced in Drosophila tissue culture cells, or in the organism, there is a concomitant increase in centrosomal MT generation (Goshima et al. 2008; Hughes et al. 2008; Wainman et al. 2009). A similar phenomenon has been reported in the absence of chromatin-driven MTs (Mottier-Pavie and Megraw 2009). As astral MTs are formed in advance of Augmin-generated ones, it is difficult to see how removal of Augmin could lead to increased centrosomal nucleation in the absence of additional regulators that recognise the overall combined contributions of the pathways. Identifying the proteins responsible for this potential cross talk should perhaps be an important goal to aid our understanding of spindle formation.
Part III: Towards an understanding of spindle formation
From the above discussion, it is clear that there is still much to learn about the way in which a cell takes a series of discrete MT-generating sites and converts them over time into an integrated, dynamically stable structure capable of both generating force and relaying positional information. It should be similarly clear that further insights will require a fusion of traditional approaches and new technologies.
Mechanistic insight is likely to come from in vitro systems in which the individual pathways can be probed. The isolation of centrosomes via centrifugation, and their subsequent immobilization onto coverslips, has been proven to be an excellent assay with which to examine centrosome-dependent MT nucleation. Initially carried out in HeLa cell extracts (Telzer and Rosenbaum 1979), similar assays have been developed in other mammalian cells (e.g. Kuriyama 1984) and Drosophila (Moritz et al. 1998).
In particular, Xenopus extracts and demembranated Xenopus sperm nuclei have allowed the development of assays to investigate centrosomal-based MT nucleation (Sawin and Mitchison 1991). Combined with immunodepletion, this has been a powerful tool for the in-depth characterisation of proteins controlling MT dynamics during spindle assembly (e.g. Boleti et al. 1996; Wittmann et al. 2000). Addition of plasmid DNA to Xenopus extracts has additionally allowed the role of chromatin-mediated MT generation to be probed (Heald et al. 1996; Sampath et al. 2004; Kelly et al. 2007). Recently, this has been taken to quantitative levels, examining the exact quantity of chromatin required to make a mitotic spindle, and the effect that chromatin shape has on the resulting spindles through lithographic micropatterning (Dinarina et al. 2009). Other advanced techniques in Xenopus, such as the use of piezoresistive dual cantilevers to quantitatively measure the mechanical forces intrinsic to in vitro spindles may, when combined with pathway disruption through immunodepletion, assist in teasing apart the properties of populations of MTs generated by different pathways (Itabashi et al. 2009).
The ability to model the dynamics of MTs, their movements in relation to one another and the activities of MAPs that regulate their function has also led to computational modelling of aspects of spindle assembly. Most notably, increasingly complex models of spindle self-organisation have been constructed based around observations from Xenopus, which predict contributions to spindle formation of MTs derived from centrosomes, chromatin and Augmin, and explain many of the emergent features and properties of a bipolar spindle (Schaffner and Jose 2006; Burbank et al. 2006; Loughlin et al. 2010). In addition, mathematical models of centrosome separation and spindle formation in Drosophila embryos have provided hypotheses that can be tested genetically or through the use of interfering antibodies (see Civelekoglu-Scholey and Scholey 2010 for review).
However, descriptive cell biology, qualitatively similar to the “poke it and see” morphological studies of old, still has much to offer, with expert observation and intuition providing insights that otherwise would go unnoticed. Moreover, by coupling cutting-edge live imaging technologies with quantification of the detailed distribution of MTs, data can be extracted and subjected to statistical analyses which can uncover trends facilitated by distinct populations of MTs over time. Qualitative observations and quantitative parameters between different samples thus allow comparative information to be obtained—relating both to the extent to which each pathway functions in different cells of the same organism (i.e. the developmental context of the pathway) and to the extent to which one pathway relates to another (though the removal or perturbation of one or more pathways in a single cell type). The main difficulty with such analyses has been the ability to image and measure the nucleation and changes in dynamics of single MTs; the density of MTs within the growing spindle is simply too great. However, there are techniques that allow such measurements to be made. The incorporation of fluorescent tubulin into non-labelled MTs, at low levels (~0.5% of total tubulin), results in the formation of MT “speckles” whose trajectories can be tracked over time (Waterman-Storer et al. 1998). This method has been used in many organisms, including newt, Xenopus, yeast and Drosophila, to investigate spindle dynamics. (Waterman-Storer et al. 1998; Maddox et al. 2000, 2002). Additionally, the ability of the MT plus end-binding protein EB1 to localise to the distal 2–3 μm of growing MTs has resulted in its use as a reporter of new MT generation—the appearance of new EB1-GFP “comets” within the mitotic spindle will occur when new MTs are actively generated. By comparing the number and dynamics of EB1-GFP punctae over time in normal cells, with those lacking particular pathways of MT generation, the contribution of these individual mechanisms has been examined (for example, Chan et al. 2005; Mahoney et al. 2006; Liang et al. 2009; Wainman et al. 2009). However, there is scope for this type of analysis to contribute much more to the understanding of MT generation during mitosis; the advent of new biophysical techniques, such as real-time super-resolution microscopy (Wolter et al. 2010) and single-molecule lifetime imaging (Mirny and Needleman 2010), coupled to faster acquisition times, should allow higher spatiotemporal resolution of the accumulation of MT nucleators simultaneously with individual MT dynamics.
Summary
The traditional view of the mitotic spindle apparatus as a molecular machine which is built through a defined irreversible set of instructions is gradually being replaced. It can instead be envisaged as a self-regulating dynamic structure where multiple pathways of MT generation are spatially and temporally controlled and integrated, constantly “talking” to one another and modifying the behaviour of their MTs in order to maintain a flexible yet robust steady-state spindle. Through taking a holistic view, methodologically and conceptually, we can continue to learn more about how this fundamental biological process takes place.
Abbreviations
- aMTOC:
-
Acentriolar microtubule organising centre
- CPC:
-
Chromosomal passenger complex
- FRET:
-
Fluorescence resonance energy transfer
- GFP:
-
Green fluorescent protein
- HAUS:
-
Homologous to Augmin subunits
- HU:
-
Hydroxyurea
- kMT:
-
Kinetochore microtubule
- MT:
-
Microtubule
- MAP:
-
Microtubule-associated protein
- MTOC:
-
Microtubule organising centre
- NEB:
-
Nuclear envelope breakdown
- PCM:
-
Pericentriolar material
- γ-TuRC:
-
γ-Tubulin ring complex
- γ-TuSC:
-
γ-Tubulin small complex
References
Andersen S, Ashford A, Tournebize R, Gavet O, Sobel A, Hyman A et al (1997) Mitotic chromatin regulates phosphorylation of Stathmin/Op18. Nature 389(6651):640–643
Bajer A, Molè-Bajer J (1986) Reorganization of microtubules in endosperm cells and cell fragments of the higher plant Haemanthus in vivo. J Cell Biol 102(1):263–281
Basto R, Lau J, Vinogradova T, Gardiol A, Woods C, Khodjakov A et al (2006) Flies without centrioles. Cell 125(7):1375–1386
Bobinnec Y, Khodjakov A, Mir L, Rieder C, Eddé B, Bornens M (1998) Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol 143(6):1575–1589
Boleti H, Karsenti E, Vernos, I (1996) Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell 84(1):49–59
Brinkley B, Zinkowski R, Mollon W, Davis F, Pisegna M, Pershouse M et al (1988) Movement and segregation of kinetochores experimentally detached from mammalian chromosomes. Nature 336(6196):251–254
Brown R, Lemmon B (1997) The quadripolar microtubule system in lower land plants. J Plant Res 110(1097):93–106
Brunet S, Polanski Z, Verlhac MH, Kubiak JZ, Maro B (1998) Bipolar meiotic spindle formation without chromatin. Curr Biol 8(22):1231–1234
Bucciarelli E, Pellacani C, Naim V, Palena A, Gatti M, Somma M (2009) Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr Biol 19:1839–1845
Burbank K, Groen A, Perlman Z, Fisher D, Mitchison T (2006) A new method reveals microtubule minus ends throughout the meiotic spindle. J Cell Biol 175(3):369–375
Calarco-Gillam P, Siebert M, Hubble R, Mitchison T, Kirschner M (1983) Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell 35(3 Pt 2):621–629
Carazo-Salas R, Guarguaglini G, Gruss O, Segref A, Karsenti E, Mattaj I (1999) Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400(6740):178–181
Casanova C, Rybina S, Yokoyama H, Karsenti E, Mattaj I (2008) Hepatoma up-regulated protein is required for chromatin-induced microtubule assembly independently of TPX2. Mol Biol Cell 19(11):4900–4908
Chan J, Calder G, Fox S, Lloyd C (2005) Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell 17(6):1737–1748
Ciciarello M, Mangiacasale R, Lavia P (2007) Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci 64(15):1891–1914
Civelekoglu-Scholey G, Scholey J (2010) Mitotic force generators and chromosome segregation. Cell Mol Life Sci 67(13):2231–2250
Colombié N, Cullen C, Brittle A, Jang J, Earnshaw W, Carmena M et al (2008) Dual roles of Incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development 135(19):3239–3246
Dinarina A, Pugieux C, Corral M, Loose M, Spatz J, Karsenti E et al (2009) Chromatin shapes the mitotic spindle. Cell 138(3):502–513
Dix C, Raff J (2007) Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol 17(20):1759–1764
Dobbelaere J, Josué F, Suijkerbuijk S, Baum B, Tapon N, Raff J (2008) A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol 6(9):e224
Gadea B, Ruderman J (2006) Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc Natl Acad Sci USA 103(12):4493–4498
Giansanti M, Bucciarelli E, Bonaccorsi S, Gatti M (2008) Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol 18(4):303–309
Gonzalez C, Saunders R, Casal J, Molina I, Carmena M, Ripoll P et al (1990) Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci 96(Pt 4):605–616
Goshima G, Mayer M, Zhang N, Stuurman N, Vale R (2008) Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181(3):421–429
Groen A, Cameron L, Coughlin M, Miyamoto D, Mitchison T, Ohi R (2004) XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr Biol 14(20):1801–1811
Gruss O, Carazo-Salas R, Schatz C, Guarguaglini G, Kast J, Wilm M et al (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104(1):83–93
Hannak E, Oegema K, Kirkham M, Gönczy P, Habermann B, Hyman A (2002) The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol 157(4):591–602
Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A et al (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382(6590):420–425
Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A (1997) Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol 138(3):615–628
Holm L, Kääriäinen S, Rosenström P, Schenkel A (2008) Searching protein structure databases with DaliLite v. 3. Bioinformatics 24(23):2780–2781
Hughes J, Meireles A, Fisher K, Garcia A, Antrobus P, Wainman A et al (2008) A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol 6(4):e98
Inoue S, Sato H (1967) Cell motility by labile association of molecules: the nature of mitotic spindle fibers and their role in chromosome movement. J Gen Physiol 50:259–292
Itabashi T, Takagi J, Shimamoto Y, Onoe H, Kuwana K, Shimoyama I et al (2009) Probing the mechanical architecture of the vertebrate meiotic spindle. Nat Meth 6(2):167–172
Kalab P, Pu R, Dasso M (1999) The ran GTPase regulates mitotic spindle assembly. Curr Biol 9(9):481–484
Kaláb P, Pralle A, Isacoff E, Heald R, Weis K (2006) Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440(7084):697–701
Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H (2007) Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell 12(1):31–43
Khodjakov A, Cole R, Oakley B, Rieder C (2000) Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 10(2):59–67
Khodjakov A, Copenagle L, Gordon M, Compton D, Kapoor T (2003) Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol 160(5):671–683
Kirschner M, Mitchison T (1986) Beyond self-assembly: from microtubules to morphogenesis. Cell 45(3):329–342
Koffa M, Casanova C, Santarella R, Köcher T, Wilm M, Mattaj I (2006) HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol 16(8):743–754
Kuriyama R (1984) Activity and stability of centrosomes in Chinese hamster ovary cells in nucleation of microtubules in vitro. J Cell Sci 66:277–295
Lawo S, Bashkurov M, Mullin M, Ferreria M, Kittler R, Habermann B et al (2009) HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol 19(10):816–826
Li H, Wirtz D, Zheng Y (2003) A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J Cell Biol 160(5):635–644
Liang Z-Y, Hallen M, Endow S (2009) Mature Drosophila meiosis I spindles comprise microtubules of mixed polarity. Curr Biol 19(2):163–168
Loughlin R, Heald R, Nédélec F (2010) A computational model predicts Xenopus meiotic spindle organization. J Cell Biol 191(7):1239–1249
Maddox PS, Bloom KS, Salmon ED (2000) The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol 2(1):36–41
Maddox P, Desai A, Oegema K, Mitchison TJ, Salmon ED (2002) Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr Biol 12(19):1670–1674
Mahoney N, Goshima G, Douglass A, Vale R (2006) Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol 16(6):564–569
Maiato H, Rieder C, Khodjakov A (2004) Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167(5):831–840
Maller J, Poccia D, Nishioka D (1976) Spindle formation and cleavage in Xenopus eggs injected with centriole containing fractions from sperm. Exp Cell Res 99(2):285–294
Maresca T, Groen A, Gatlin J, Ohi R, Mitchison T, Salmon E (2009) Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol 19(14):1210–1215
Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW (2004) The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165(5):673–683
Mastronarde D, McDonald K, Ding R, McIntosh J (1993) Interpolar spindle microtubules in PTK cells. J Cell Biol 123(6 Pt 1):1475–1489
Matthies H, McDonald H, Goldstein L, Theurkauf W (1996) Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol 134(2):455–464
McGill M, Brinkley BR (1975) Human chromosomes and centrioles as nucleating sites for the in vitro assembly of microtubules from bovine brain tubulin. J Cell Biol 67(1):189–199
Megraw T, Kao L, Kaufman T (2001) Zygotic development without functional mitotic centrosomes. Curr Biol 11(2):116–120
Meireles A, Fisher K, Colombié N, Wakefield J, Ohkura H (2009) Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184(6):777–784
Merdes A, Ramyar K, Vechio J, Cleveland D (1996) A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87(3):447–458
Mirny L, Needleman D (2010) Quantitative characterization of filament dynamics by single-molecule lifetime measurements. Meth Cell Biol 95:583–600
Moore W, Zhang C, Clarke P (2002) Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr Biol 12(16):1442–1447
Moritz M, Zheng Y, Alberts B, Oegema K (1998) Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol 142(3):775–786
Mottier-Pavie V, Megraw T (2009) Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell 20(10):2605–2614
Nicklas RB, Ward SC (1994) Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol 126(5):1241–1253
O'Connell CB, Loncarek J, Hergert P, Kourtidis A, Conklin DS, Khodjakov A (2008) The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J Cell Biol 183(1):29–36
O'Connell C, Loncarek J, Kaláb P, Khodjakov A (2009) Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J Cell Biol 187(1):43–51
Rebollo E, Llamazares S, Reina J, Gonzalez C (2004) Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol 2(1):E8
Rieder C (2005) Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 114(5):310–318
Rogers G, Rusan N, Peifer M, Rogers S (2008) A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell 19(7):3163–3178
Ruchaud S, Carmena M, Earnshaw W (2007) Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 8(10):798–812
Sampath S, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H (2004) The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118(2):187–202
Sawin K, Mitchison T (1991) Mitotic spindle assembly by two different pathways in vitro. J Cell Biol 112(5):925–940
Schaffner S, José J (2006) Biophysical model of self-organized spindle formation patterns without centrosomes and kinetochores. Proc Natl Acad Sci USA 103(30):11166–11171
Schatz C, Santarella R, Hoenger A, Karsenti E, Mattaj I, Gruss O et al (2003) Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J 22(9):2060–2070
Schuh M, Ellenberg J (2007) Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130(3):484–498
Shimamura M, Brown R, Lemmon B, Akashi T, Mizuno K, Nishihara N et al (2004) Gamma-tubulin in basal land plants: characterization, localization, and implication in the evolution of acentriolar microtubule organizing centers. Plant Cell 16(1):45–59
Smirnova E, Bajer A (1994) Microtubule converging centers and reorganization of the interphase cytoskeleton and the mitotic spindle in higher plant Haemanthus. Cell Motil Cytoskeleton 27(3):219–233
Snyder JA, McIntosh JR (1975) Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. J Cell Biol 67(3):744–760
Sonbuchner TM, Rath U, Sharp DJ (2010) KL1 is a novel microtubule severing enzyme that regulates mitotic spindle architecture. Cell Cycle 9(12):2403–2411
Srayko M, O'toole E, Hyman A, Müller-Reichert T (2006) Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol 16(19):1944–1949
Steffen W, Fuge H, Dietz R, Bastmeyer M, Müller G (1986) Aster-free spindle poles in insect spermatocytes: evidence for chromosome-induced spindle formation? J Cell Biol 102(5):1679–1687
Teixidó-Travesa N, Villén J, Lacasa C, Bertran M, Archinti M, Gygi S et al (2010) The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol Biol Cell 21(22):3963–3972
Telzer B, Rosenbaum J (1979) Cell cycle-dependent, in vitro assembly of microtubules onto pericentriolar material of HeLa cells. J Cell Biol 81(3):484–497
Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F (2008) Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell 19(5):1873–1882
Trieselmann N, Armstrong S, Rauw J, Wilde A (2003) Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J Cell Sci 116(Pt 23):4791–4798
Tseng B, Tan L, Kapoor T, Funabiki H (2010) Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell 18(6):903–912
Tulu U, Fagerstrom C, Ferenz N, Wadsworth P (2006) Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol 16(5):536–541
Uehara R, Nozawa R-s, Tomioka A, Petry S, Vale R, Obuse C et al (2009) The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci USA 106(17):6998–7003
Varmark H (2004) Functional role of centrosomes in spindle assembly and organization. J Cell Biochem 91(5):904–914
Wainman A, Buster D, Duncan T, Metz J, Ma A, Sharp D et al (2009) A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev 23(16):1876–1881
Wakefield JG, Huang J-Y, Raff J (2000) Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr Biol 10(21):1367–1370
Wakefield JG, Bonaccorsi S, Gatti M (2001) The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol 153(4):637–648
Waterman-Storer C, Desai A, Bulinski J, Salmon E (1998) Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol 8(22):1227–1230
Wittmann T, Wilm M, Karsenti E, Vernos I (2000) TPX2, a novel Xenopus MAP involved in spindle pole organization. J Cell Biol 149(7):1405–1418
Wolter S, Schüttpelz M, Tscherepanow M, VAN DE Linde S, Heilemann M, Sauer M (2010) Real-time computation of subdiffraction-resolution fluorescence images. J Microsc 237(1):12–22
Wong J, Fang G (2006) HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol 173(6):879–891
Yang C-P, Fan S-S (2008) Drosophila mars is required for organizing kinetochore microtubules during mitosis. Exp Cell Res 314(17):3209–3220
Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj I, Karsenti E (2009) ISWI is a RanGTP-dependent MAP required for chromosome segregation. J Cell Biol 187(6):813–829
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinform 9:40–43
Zhu H, Coppinger J, Jang C-Y, Yates J, Fang G (2008) FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol 183(5):835–848
Zimmerman W, Sillibourne J, Rosa J, Doxsey S (2004) Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 15(8):3642–3657
Acknowledgements
We would like to thank Yaseen Ladak for initiating the structural alignments of Augmin subunits, Charlotte Deane for advice, the Wakefield lab for stimulating discussion and the referees for helpful and insightful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: James Wakefield and Herbert Macgregor
Rights and permissions
About this article
Cite this article
Duncan, T., Wakefield, J.G. 50 ways to build a spindle: the complexity of microtubule generation during mitosis. Chromosome Res 19, 321–333 (2011). https://doi.org/10.1007/s10577-011-9205-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9205-8