Abstract
The notion of nucleic acids in the spindle, and particularly, the centrosome has a long history of inquiry, doubt, and debate. However, the association of specific RNAs with these structures is now confirmed by several investigators. What their presence means for the assembly, function, and evolution of the cell division apparatus is not known; but with newly available information and probes, these are questions that can finally be addressed. The present article summarizes the history of this field, what we know about the molecules in question, and in light of these findings, emphasizes the need to view the cell division apparatus for what it is by definition, a ribonucleoprotein complex.
Similar content being viewed by others
The subject in context
The presence of nucleic acids in centrosomes and the spindle have been proposed, observed, and reported since the 1950s. Why did the subject remain, perhaps even until today, such a controversial issue? The explanation is manifold and includes legitimate concern over contamination from other cellular compartments in biochemical preparations. With a typically high background of cytoplasmic ribosomes, even microscopic images of stained intact cells could be difficult to interpret. Also, evidence for RNA and DNA in centrosomes accumulated for approximately 40 years but was interspersed with contradictory studies, primarily regarding the presence of DNA (reviewed in Johnson and Rosenbaum 1991; Marshall and Rosenbaum 2000). Perhaps less tangible but still a likely cause for lingering controversy is that the presence of nucleic acids in the spindle or centrosomes will require us to look differently at these structures from a functional, and more to the point, evolutionary standpoint.
From the earliest studies, our overriding focus has been on the protein components of the cell division apparatus. The centrosome, centriole, and particularly the spindle, were long thought of as, first and foremost, microtubule-based structures. After all, microtubules are the most visually dominant structures, at least with the imaging techniques and probes we have had until recently, and tubulin is the dominant protein component. As other protein components were discovered, the analytical frame of reference was usually their protein–protein interactions with tubulin. Nucleic acids, in the form of chromatin, were cargo (albeit with a more proactive role, we learned as time passed). However, there is evidence that all four classes of biomolecules—protein, carbohydrate (Chang et al. 2004), lipid (Ackerman 1961; Tsai et al. 2006), and nucleic acid (references below)—are closely associated with the spindle and integral to its structure and function. We are likely selling this biological machine short by referring to it only as a ribonucleoprotein complex (RNP). These other classes of biomolecules must be incorporated into our models for the genesis and function of the centrosomes and spindle, especially since their presence and activities may precede those of the hallmark proteins we have focused on for so long.
Thinking of the cell division apparatus as not just a protein machine, but as a comprehensive unit composed of all classes of biomolecules (almost a cell within a cell) may also help us to understand its evolution, and by extension, eukaryosis. We agree that approaching the cell division apparatus as an evolutionary seme (Margulis et al. 2007) will ultimately be more informative than focusing on individual (or small groups of) molecules. This can make the difference, for example, between viewing nucleic acids or lipids and their respective metabolizing enzymes as later recruits rather than original components. A step in this direction is to broaden our description of the cell division apparatus to that of a ribonucleoprotein complex. By definition, it is.
Experimental background
Early efforts to define the molecular composition of the mitotic apparatus by Amano (1954), Stich (1954), Mazia (1955), Shimamura and Ota (1956), Rustad (1959), Zimmerman (1960), and others revealed evidence for RNA. These studies had several limitations that have been largely overcome in later years due to the development of higher resolution imaging and more specific histochemical probes. The early analyses were also subject to a ubiquitous problem faced even today: when the cell and its compartments are disrupted, molecules that do not normally associate in situ may adsorb to each other artifactually. Nonetheless, these early studies, no different from contemporary work successfully defining the composition of membranes, mitochondria, and other cellular compartments, paved the way for more refined dissection of the spindle and associated structures. The evidence for RNA in the cell division apparatus can be divided into three categories, morphological, functional, and accessory, as summarized below. A number of published results are not described here because potential problems with labeling specificity or cell fractionation artifact make them more difficult to interpret with confidence. However, others withstand reasonable scrutiny, in part or in whole.
Morphological evidence for centrosomal RNA
There are a number of published papers aiming to show the association of nucleic acids with centrioles, centrosomes, or basal bodies by histochemical localization. One of the earliest of these studies was Hartman et al. (1974), who used the combined approaches of cell fractionation, acridine orange fluorescence, and nucleic acid biochemistry to provide evidence for the presence of RNA in the basal bodies of Tetrahymena pyriformis. Cell pellicles (a complex of the cell surface and underlying cortex) were isolated and observed by electron microscopy to assess their content and structure. Prominent basal bodies and little other cytoplasmic structure were visible. When pellicles were stained with acridine orange, they fluoresced orange in a punctate pattern corresponding to the basal body pattern. Acridine orange is a nucleic acid selective dye that fluoresces green in the presence of DNA and red when bound to RNA, so the color of the fluorescence here indicated a significant RNA component. In support of this, acridine fluorescence was dissipated with RNase, but not DNase treatment. That the fluorescent nucleic acid signal was due to the presence of RNA was confirmed by the preferential incorporation of 3H-uridine vs. 3H-thymidine in metabolic labeling experiments. The authors made a significant effort using sedimentation and hybridization analysis to account for potential contamination by cytoplasmic mRNA, tRNA, or rRNA. This study was among the most comprehensive of its contemporaries but, since all data were obtained from disrupted cells, the possibility still exists that the results, including the acridine localization, were due to cytoplasmic contamination.
In a later study, Dippel (1976) performed a careful ultrastructural analysis of basal bodies in intact Paramecium and then observed the effects of RNase, DNase, and protease on that structure. The author described a luminal complex within the core of the basal body, a “twisted or looped 90 Å diameter fiber, or more probably pair of fibers in association with dense granules.” Prolonged DNase treatment had no detectable effect on basal body structure, particularly in reference to the luminal complex. Pronase treatment, as would be expected for a largely proteinaceous structure, had a series of time-dependent effects. RNase preferentially and completely dissolved the luminal complex. It is possible that the commercial RNase preparation used in these experiments was contaminated with proteolytic enzymes, particularly since the luminal complex was affected by pronase treatment as well (although differentially). However, the more likely explanation, and coincidentally the simplest, is that the luminal complex is an RNP, and therefore, susceptible to hydrolysis by either enzyme. The contamination hypothesis also requires the assumption that the RNase preparation was selectively compromised with protease, but not the DNase.
Rieder subsequently (1979) used Bernhard’s method of uranyl staining followed by EDTA bleaching (Bernhard 1969) to visualize RNP complexes at the ultrastructural level in newt lung cells. Organelles and molecular assemblies in the same thin section known a priori to either lack or contain RNA served as controls for both histochemical staining and its subsequent abolition. The results were that ribosomes, kinetochores, and centrioles stained as RNPs. In centrioles, RNP staining was found on the inner surface of the centriole triplet blades and on the foot of the A tubule. This pattern was thoroughly abolished by RNase treatment. Alternative explanations for these results are that the RNase preparation contained protease contamination or that Bernhard’s RNP method is not adequately specific. However, given the in situ positive and negative controls as well as other analyses described in the paper, the best and simplest explanation is, again, that centrioles contain RNA.
Finally, in an innovative but little known study, Laane and Haugli (1974) present intriguing evidence for the association of RNA with centrosomes in the slime mold, Physarum. The authors used acridine orange to follow the formation of division centers during the centrioless, intranuclear mitosis characteristic of the Physarum plasmodium. A microscope modified to enhance red/green fluorescence sensitivity and differentiation was used to observe the process in real time. The authors describe the emergence of a single RNA-containing particle from the nucleolus and, using a combination of acridine orange fluorescence and brightfield techniques, describe the division of this aggregate into two and the appearance of two foci at the spindle poles. Although the authors indicate their confidence in this sequence of events based on the observation of many cells, difficulties remain in equating the acridine orange-foci with the phase-dense foci at the spindle poles. However, some very early studies (Lavdowsky 1894) as well as recent ones (Alliegro et al. 2010) are consistent with this overall theme, suggesting that Laane and Haugli’s observations represent the first view of centrosomes imaged in real time via their RNA content.
Functional studies
Physiological studies, for the most part, apply the same rationale as the morphological studies described above: demonstrating RNase sensitivity of certain centrosome-based activities. Evidence for the presence of RNA in basal bodies and a possible role in microtubule nucleation was described by Hiedemann et al. (1977). Isolated basal bodies, when injected into Xenopus oocytes, induce the formation of asters. Heidemann et al. observed that RNase treatment of basal bodies before injection eliminated their aster-forming potential. These results were not produced with DNase treatment and suggest that RNA present in basal bodies plays a role in either capturing or nucleating microtubules. A criticism of these experiments is that injected basal bodies can cluster, and such aggregates can artifactually induce aster formation (Marshall and Rosenbaum 2000). Asters formed in this way may, therefore, be physiologically irrelevant, rendering the results of RNase treatment likewise. It has also been suggested, since certain basic proteins are known to promote microtubule growth in vitro, that the removal of basic ribonucleoproteins (perhaps contributed by contaminating ribosomes) by RNase treatment could remove an aster-forming activity not attributable specifically to RNA (Johnson and Rosenbaum 1991). As a general counterpoint to this latter criticism, it can be argued that if bound RNPs are able to constitute an RNase-sensitive aster-forming activity in vitro, then, given the soup of RNPs in which the spindle exists in situ, they could indeed constitute an aster-forming complex that is relevant in the living cell.
In any case, observations of other investigators using lysed cell systems mitigate against some of these potential weaknesses. For example, it was shown that RNase T1 and RNase A both degrade pericentriolar material in lysed PtK2 cell preparations with a concomitant loss in microtubule nucleation potential (Pepper and Brinkley 1980). Thus, a clear alteration in centrosome morphology was correlated with a physiological effect. Neither of these two enzymes altered the structure or microtubule nucleating activity of kinetochores. Conversely, DNase 1 affected both the structure and nucleating activity of kinetochores, but had no effect on centrosomes. In a similar study, Snyder (1980) showed that RNase A or T2, but not DNase 1, inhibited microtubule nucleation in PtK1 cells.
There are a number of other reports implicating RNA in centrosome function. Zackroff et al. (1976) demonstrated the effects of RNase on aster symmetry and fiber length and was able to reverse the observed effects with the RNase inhibitor polyguanylic acid. Peterson and Berns (1978) inhibited spindle formation using the light-activated nucleic acid binding dyes, psoralens. In these studies, PtK2 cells were treated with psoralens of varying affinity, followed by laser microbeam targeting of the centriolar region. Psoralens selective for DNA had no effect, while those of broader specificity—for both RNA and DNA—inhibited spindle formation.
Related observations
There are at least two other reports that, although they do not demonstrate spindle- or centrosome-associated RNA, are nonetheless intriguing. Nor was the existence of centrosome- or spindle-associated nucleic acids the focus of these studies. Nevertheless, the observations take on additional interest in the context of those described above. Both of the studies here summarized are related to enzymes involved in nucleic acid metabolism.
Free purines are salvaged for reuse in nucleotide and nucleic acid synthesis by two major mechanisms, the primary one being mediated by the action of purine phosphoribosyltransferase. Another important pathway is that mediated by purine nucleoside phosphorylase (PNP). Oliver et al. (1981) have shown by enzyme histochemistry and immunohistochemistry that PNP is present at the site of centrioles and basal bodies in several mammalian and protist cells. The function of the enzyme at this site has not been determined and could range from the metabolism of guanosine derived from localized GTP hydrolysis, to the localized synthesis of RNAs involved in, for example, centrosome assembly. For the latter possibility, a localized source of nucleic acid precursors via a PNP-mediated salvage pathway would be of little use without similarly localized synthetic machinery, including RNA polymerase. And so another intriguing observation is the localization of RNA polymerase II to the centrosomes and spindle (Wulf et al. 1980). In this report, RNA polymerase II was localized using a fluorescent α-amanitin conjugate. α-Amanitin interacts with the RNA polymerase II bridge helix and retards progression of the polymerase. Inhibition is therefore closely related to the interaction between RNA and its DNA template. An antibody to polymerase II protein showed overlapping but much broader distribution, so alternative explanations are possible, although the authors made a thorough job of characterizing the specificity of their fluorescent α-amanitin probe. The purpose for these two enzymes (that are normally involved in nucleic acid metabolism) concentrating at the spindle pole is not clear. However, our new knowledge that centrosomes and the spindle are RNPs may bring new meaning to the observations.
Findings of specific nucleic acids associated with the centrosome and spindle
The evidence for DNA in centrioles and centrosomes has been met in almost every case with contradictory observations, so that to this day there is no consensus for its existence (reviewed in: Johnson and Rosenbaum 1991; Marshall and Rosenbaum 2000). There were, however, two key differences in the search for RNA associated with the cell division apparatus. First is that considerably more evidence was accumulated; biochemical and morphological at the light and electron microscopic levels. Moreover, pitfalls and alternative explanations could be found for most individual observations, but they were often more complicated and no more likely than the original, positive interpretation. Second, and far more important, is that evidence for specific centrosomal and spindle RNAs was ultimately uncovered. The data has been registered by at least five independent laboratories, as described in the following sections. The issue of whether RNA is associated with the centrosome and spindle is, therefore, no longer in question. The status of these molecules as transient or residential, and the functional and evolutionary implications, certainly are.
For purposes of discussion, these implications can be divided into four areas. The first, that RNA is targeted to the centrosome and spindle to effect localized translation of proteins involved in cell division, is not controversial. It is not yet proven, but it is reasonable and has precedent in other areas. The second, that RNA is present transiently in the centrosome for purposes of trafficking via microtubules, is supported by good evidence. Third, the suggestion that RNA plays a structural role in the assembly or organization of the centrosomes and spindle is also unproven but not unlikely. Last is the idea that centrosome- and/or spindle-associated RNAs are derived consequences of a symbiogenetic event. This remains speculative, although there are clues to suggest it is possible. It is controversial and is sometimes met with derision, as was the general concept of symbiogenesis itself at one time. Regardless, it is important to note that none of these four possibilities are mutually exclusive and that any combination of them may be true.
The first evidence indicating the localization of specific RNAs at the spindle and spindle pole was described by Groisman et al. (2000). This report echoes the earlier studies of Mazia and others; however, Groismann et al. had at their disposal the molecular probes to permit identification of two specific transcripts involved. The study has at its center the observation that translational control of certain proteins in the embryo is mediated by cytoplasmic polyadenlylation. Xenopus Bub3 and cyclin B1 mRNAs contain the necessary cis-acting sequences mediating cytoplasmic polyadenylation, and the proteins (CPEB and maskin, among them) that bind to these regulatory sites localize to the spindle. Groisman et al. (2000) correctly hypothesized a similar distribution for the target mRNAs, and indeed found Bub3 and cyclin B1 mRNAs localized to the spindle and spindle pole. The authors propose that the mRNAs of these cell cycle regulators are targeted to the spindle to precisely control translation both temporally and spatially. This scenario has ample precedent (Gavis et al. 2007) and is probably the case for certain centrosome- and spindle-associated RNAs, but there are other groups of transcripts that are not likely being utilized in this manner, and these fall into two classes.
The first class includes RNAs that are present on microtubules and in association with the centrosome for purposes of trafficking. These are transients, en route to localization elsewhere in the cell or to specific blastomeres in the embryo, as has been demonstrated in the snail, Ilyanassa (Lambert and Nagy 2002). Transcripts of the developmental patterning genes, even-skipped (Eve), decapentaplegic (Dpp), and tolloid (Tld) are distributed diffusely in the cytoplasm of all blastomeres at the four cell stage. They become localized to the centrosome during interphase in a microtubule-dependent manner and are then transported to a region of the blastomere cortex inherited by only one of the two daughter cells during prophase of the third cleavage division. Eve, Dpp, and Tld are all well-characterized molecules. They were defined and known for years as cytoplasmic transcripts, so it is clear a priori that they do not represent a set of RNAs intrinsic to the centrosome. Nevertheless, the study demonstrates a class of centrosome-associated RNAs, even if the association is transient and exemplifies an extrinsic function. A subsequent screen for centrosome-associated RNAs in Ilyanassa embryos resulted in the identification of approximately 50 additional molecules considered highly enriched (Kingsley et al. 2007). A theme common to this and other studies (Alliegro et al. 2006; Blower et al. 2007; Lécuyer et al. 2007; Alliegro and Alliegro 2008) is a preponderance of genes unmatched in databases as well as an overrepresentation of genes involved in DNA and RNA metabolism.
Although studies by Blower et al. (2005; 2007) did not involve the localization of specific sequences in situ, they provide another example of centrosome- and spindle-associated RNAs. Rae1 is an mRNA export protein shown to localize to the spindle and demonstrated to exist as part of an RNP complex in cell extracts. This Rae1-RNA complex was found to be required for spindle assembly. Functional studies involved treating Rae1-RNA complexes with RNase A to abolish spindle-forming activity, but took advantage of one additional tool not used by investigators in the 1960s and 1970s to support the hypothesis that spindle-forming activity was specifically RNase-sensitive; RNase A proteolytic fragments that are, by themselves, inactive, but that can be combined to reconstitute enzymatic activity. Addition of the inactivated RNase A or its proteolytic fragment did not abolish spindle-forming activity of Rae1 RNP, but addition of the two fragments together did. Thus, Rae1-RNP localizes to the spindle and requires its RNA component to function in spindle assembly. In support of the earlier Xenopus study (Groisman et al. 2000), Blower et al. (2007) provide evidence for localized translation using a fluorescent analogue of puromycin in Xenopus egg extracts. The authors conclude that this represents a conserved and widely used mechanism for enhancing protein localization, in this case proteins involved in cell cycle regulation. However, several observations suggest there may be more to this story. First is that the majority of spindle-associated RNAs in this study are not translated, nor is translation required for their localization to the cell division apparatus. Unlike Bub3 and cyclin B1 mRNAs, 93% of the transcripts identified in this report (Blower et al. 2007) do not contain cytoplasmic polyadenylation regulatory sequences, and a significant proportion, as described for other studies, are either unidentifiable in databases or represent genes involved in nucleic acid metabolism. Moreover, a dysmorphic spindle is induced by Rae1 depletion as well as RNase treatment of cell extracts. It therefore appears that the association of at least some of these transcripts with the spindle and spindle pole has less to do with localized translation and perhaps more to do with organization and assembly of the cell division apparatus.
This possibility is discussed more fully in a report by Lécuyer et al. (2007). The authors describe the localization of specific RNAs to various cellular domains in Drosophila embryos by in situ hybridization, including the plasma membrane, cell junctions, and the nucleus. The study is not focused specifically on the cell division apparatus, but evidence is presented for 33 specific RNAs that are so localized. Straightforward statistics were used to estimate a total of 134 such RNAs, genome-wide. Of these, six were directly localized to the centrosome (estimated 24, genome-wide) with ten more described as microtubule-associated, ten as spindle-midzone-, and 14 chromatin-associated. This study does not address the function of spindle- and centrosome-associated RNAs experimentally, but the authors reason that since RNA localization usually precedes the localization of their encoded proteins, and since this pattern is so pervasive, it is likely that these transcripts play a role in the nucleation and assembly of the protein complexes in question. They also point out that similarly localized non-coding RNAs are known to play a structural or catalytic role independent of translation.
Three other studies have found specific RNAs associated with the centrosomes and/or spindle in surf clam (Spisula solidissima) oocytes (Alliegro et al. 2006; Alliegro and Alliegro 2008; Alliegro et al. 2010). The first two reports described a set of RNAs that are significantly enriched in isolated centrosomes and includes confirmatory in situ hybridizations for four of these. These four do not appear to be localized within the centrosome; rather, the centrosome appears to be localized within the RNA hybridization patch (Fig. 1). However, this is not conclusive because the resolution of light microscopic in situ hybridization may be deceiving, especially since the colored product of the phosphatase reaction can diffuse to some extent. Also, localization of more than 20 other candidate transcripts has yet to be done. It is therefore unclear at this time whether any of these RNAs are preferentially associated with the centriole or pericentriolar matrix.
Newly formed centrosomes embedded within a patch of RNA. a In situ hybridization showing the distribution of cnRNA 239 (arrowhead) in a parthenogenetically activated (4-min) surf clam oocyte. b Immunofluorescent localization of centrosomes using an antibody to γ-tubulin. Two distinct γ-tubulin foci can be resolved (arrows). c Overlay of a and b illustrating the embedment of the two centrosomes within the cnRNA 239 hybridization patch. Size bar = 15 μm
As with the studies of Blower et al. (2007), Kingsley et al. (2007), and Lécuyer et al. (2007), a significant percentage of these Spisula centrosome-associated RNAs (cnRNAs) have not been identified. Another unique quality of this set of RNAs is that they are exceedingly intron-poor, which stands in contrast to Tld, Eve, Dpp, Bub3, cyclin B1, and the others named above, all of which have an intron content comparable with the balance of their respective genomes. It is possible that the contrasting gene structures delineate distinct sets of centrosome and spindle-associated RNAs; one set transient and targeted to, or through the centrosome, and a second set that may be best described as a “resident transcriptome.”
Recently, a character that could shed some light on questions of centrosome content, biogenesis, and evolution has returned to the stage: the nucleolinus (Alliegro et al. 2010). Although most readers will not be familiar with this structure, it is by no means a “new invention.” Its history can be traced back at least 150 years from Agassiz (1857), through Montgomery (1898), Carleton (1920), Allen (1951; 1953), and Love and colleagues (Love and Bharadwaj 1959; Love and Liles 1959; Love and Wildy 1963; Love 1965, 1966; Love and Walsh 1968; Love and Soriano 1971). Still, the literature on the nucleolinus is scanty and we know little about it, other than to say it is an RNA-rich compartment closely associated with the nucleolus in a spectrum of cell types. In some cells, such as Spisula oocytes, it is a distinct morphological entity. In other cells, it appears to be integrated within the nucleolus, and histological stains that are no longer in common use are required for its visualization (Alliegro 2011).
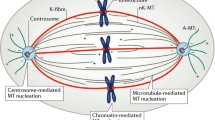
The nucleolinus is brought into this discussion because it was proposed to play a spindle-forming role in Spisula oocytes entering meiosis (Allen 1951; 1953) and was long ago thought to be a direct precursor of centrosomes (Lavdowsky 1894). Moreover, riboprobes to nucleolinar RNAs reveal an association with the centrosomes and spindle suggestive of a preexisting matrix, and experimental evidence supports the case for a role of the nucleolinus in forming the cell division apparatus (Fig. 2). Induction of damage to the nucleolinus via laser microsurgery before centrosomes or a spindle form results in a dysmporhic meiotic spindle and failed chromatin segregation in parthenogenetically activated oocytes (Alliegro et al. 2010). Centrosomes can form, but their size and number appear to be disregulated. In oocytes fertilized with sperm after microsurgery, mitotic centrosomes and the spindle fail to form altogether. The precise role of the nucleolinus in these processes is not yet known, due in part to the complex origin of the microtuble organizing center during the first three division cycles in these zygotes. Spisula oocytes are arrested in prophase I of meiosis. The first two (meiotic) divisions, which can be initiated by fertilization or parthenogenetic activation, are organized by maternally derived centrosomes. The first centrosome is generated “de novo.” It is not clear if the second centrosome is formed by duplication of the first, or if both arise simultaneosly. At the end of meiosis, the maternal centrosomes are suppressed and mitosis is governed by centrosomes derived, at least in part, from sperm (Wu and Palazzo 1999). The mitotic centrosomes are replication-competent and carry the developing embryo through many rounds of cell division. Regardless of the chain of events at the molecular level, it is clear that nucleolinar RNAs are closely associated with the centrosome and spindles during the early stages of meiosis and that this RNP is important for the formation of a normal, functioning cell division apparatus. These observations are reminiscent of Laane and Haugli’s (1974) and may underlie experimental results of 50 years ago using laser ablation (Gaulden and Perry 1958) as well as more recent ones obtained with the benefit of newer molecular tools (Ugrinova et al. 2007) showing that the nucleolus is directly involved in cell division. The nucleolinus may represent that domain of the nucleolus responsible for its cell cycle-related functions. In some cell types, such as oocytes, the functional unit is gathered into a discrete morphological unit; in other cells, it is in a more integrated state.
Centrosome formation in oocytes of the surf clam (Spisula) from the nucleolinar RNP. Unfertilized oocytes of Spisula solidissima are arrested in prophase of meiosis I. A large tetraploid nucleus, or germinal vesicle (GV) can be seen, with chromosomes (blue) attached to the nuclear envelope. Within the GV is a prominent, nearly spherical nucleolus (gray circle within the GV) and nucleolinus (red circle). Centrosomes are not present in the unfertilized oocyte, they are formed “de novo” approximately 4 min after fertilization. Likewise, most centrosomal RNAs are not expressed before fertilization. However, some are present in the unactivated oocyte, in the nucleolinus. Within the first few minutes of fertilization, elements of the RNA-rich nucleolinus “bud” from the structure. These procentrosomes recruit γ-tubulin at approximately 4–5 min (now shown in green in the cytoplasm of the 8-min zygote) and are recognizable as centrosomes. The GV and nucleolus dissipate during this time span. Shortly thereafter, the nucleolinus is no longer visible as a distinct structure, but its components can be visualized by in situ hybridization using probes to nucleolinar RNA (red). The centrosomes, chromosomes, and developing spindle come to lie in a matrix of RNA (Alliegro et al. 2010 and M.C. Alliegro and M.A. Alliegro, unpublished observations)
Implications
The idea of nucleic acids associated with the spindle, and especially, the centrosome (centrioles/basal bodies), elicited a great deal of excitement over one-half century ago and then entered a period of harsh skepticism for well over a decade. Today, unless we choose to discount the technique of in situ RNA hybridization altogether, the evidence for specific RNAs associated with the spindle and centrosomes is overwhelming. At least 15 distinct molecules for which the full length sequences are known have been localized to the centrosome by in situ hybridization in four laboratories (Lambert and Nagy 2002; Alliegro et al. 2006; Alliegro and Alliegro 2008; Kingsley et al. 2007; Lécuyer et al. 2007). Others have been localized to the spindle (Groisman et al. 2000; Lécuyer et al. 2007; Alliegro et al. 2010) and dozens more are either presumed (by biochemical enrichment) or known (by in situ localization based on partial sequences) to be present in these structures (Lécuyer et al. 2007; Alliegro and Alliegro 2008). We may now return our attention to the function and origin of spindle and centrosomal RNAs.
That RNAs of differentiation factors can be routed through centrosomes for delivery to select embryonic cells or that others can be directed to specific cytoplasmic domains for localized translation are, by now, unsurprising. We have much to learn about these important processes. Yet, there are at least two other areas of inquiry regarding centrosome- and spindle-associated RNA with broader implications for cell biology: (1) the role of these nucleic acids in the assembly and organization of the microtubule-based cytoskeleton and (2) the origin and evolution of these RNAs and the structures with which they are associated.
These two questions regarding function and evolution, respectively, encompass key elements of the original paradigm that elicited so much interest. Centriole replication has been described as generative, semiconservative, and independent of the nucleus. Based on these observations, it was posited that the duplication process could be templated by nucleic acid. It followed that the presence of nucleic acids in the centriole or centrosome could signify an exogenous evolutionary origin, a model fueled by discoveries concerning mitochondria and chloroplasts. Recent experimental evidence (Loncarek et al. 2007) and the benefits of hindsight suggest that the first (functional) component of the paradigm should be modified: nucleic acids template nucleic acids, though probably not organelles. However, nucleic acids do serve to scaffold macromolecular assemblages, as has been shown for RNA in the assembly and organization of the ribosome (Woodson 2008) and the bacterial nucleoid (Pettijohn and Hecht 1974). At the organellar level, the nucleus is ultimately scaffolded upon nucleic acids (chromosomes) at the conclusion of each telophase. Examples such as these leave little doubt that nucleic acids do play a role in the assembly of the centrosome and spindle, though perhaps not by the mechanism of direct templating originally hypothesized.
The second half of the paradigm, that these RNAs are remnants of an organellar genome, was mistakenly dismissed over our lack of definitive evidence for their existence from the 1970s until recently. Though this question remains unanswered, it should be the subject of reinvigorated inquiry as a result of recent findings. There is currently no evidence to directly support or refute this hypothesis, but there are some tantalizing observations in its favor. These include the intronless structure of at least some cnRNA genes (Alliegro et al. 2010) and their relationship to viral sequences (Alliegro and Satir 2009). Findings of function, whether for trafficking or localized translation, have no bearing on the question, since identifying one function does not preclude another, much less an evolutionary history. Moreover, any organelle or molecular assemblage may be adopted for—so it may seem to us—unexpected or even disparate functions. The nucleolus’ long-recognized function in ribosome biogenesis vs. its relatively unexplored role in cell cycle progression serves as a keen example of this principle. The hypothesis that centrosomal (or spindle) RNAs are descendants of an ancient endosymbiont remains as intriguing, provocative, and possible as ever. Meanwhile, our understanding of these structures as ribonucleoprotein complexes should broaden our grasp of their function and evolution as we uncover the rest of this story.
Abbreviations
- Eve :
-
Even-skipped
- cnRNA:
-
Centrosome-associated RNA
- CPEB:
-
Cytoplasmic polyadenylation element binding protein
- Dpp :
-
Decapentaplegic
- GV:
-
Germinal vesicle
- PNP:
-
Purine nucleoside phosphorylase
- RNP:
-
Ribonucleoprotein
- Tld :
-
Tolloid
References
Ackerman GA (1961) Histochemistry of the centrioles and centrosomes of the leukemic cells from human myeloblastic leukemia. J Biochem Biophys Cytol 11:717–719
Agassiz L (1857) Contributions to the natural history of the United States of America. First monograph. Part IIL Embryology of the Turtle, Boston
Allen RD (1951) The role of the nucleolus in spindle formation. Biol Bull 101:214
Allen RD (1953) Fertilization and artificial activation in the egg of the surf calm, Spisula solidissima. Biol Bull 105:213–239
Alliegro MC (2011) The nucleolinus: a disappearing, forgotten, and (maybe) misnamed organelle. Comm Int Biol (in press).
Alliegro MC, Alliegro MA (2008) Centrosomal RNA correlates with intron-poor nuclear genes in Spisula oocytes. Proc Natl Acad Sci USA 105:6993–6997
Alliegro MC, Satir P (2009) Origin of the cilium: novel approaches to examine a centriolar evolution hypothesis. Meth Cell Biol 94:51–63
Alliegro MC, Alliegro MA, Palazzo RE (2006) Centrosome-associated RNA in surf clam oocytes. Proc Natl Acad Sci USA 103:9034–9038
Alliegro MA, Henry J, Alliegro MC (2010) Rediscovery of the nucleolinus, a dynamic RNA-rich organelle associated with the nucleolus, spindle, and centrosomes. Proc Natl Acad Sci USA 107:13718–13723
Amano S (1954) Structure and function of the central body and the nucleolus. Acta Med Univ Kyoto 37:5–27
Bernhard W (1969) A new staining procedure for electron microscopical cytology. J Ultrastruct Res 27:250–265
Blower MD, Nachury M, Heald R, Weis K (2005) A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121:223–345
Blower MD, Feric E, Weis K, Heald R (2007) Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol 179:1365–1373
Carleton HM (1920) Observations on an intra-nucleolar body in columnar epithelial cells of the intestine. Quart J Microsc Sci S2-64:329–341
Chang P, Jacobson MK, Mitchison TJ (2004) Poly(ADP-ribose) is required for spindle assembly and structure. Nature 432:645–649
Dippel RV (1976) Effects of nuclease and protease digestion on the ultrastructure of Paramecium basal bodies. J Cell Biol 69:622–637
Gaulden ME, Perry RP (1958) Influence of the nucleolus on mitosis as revealed by ultraviolet microbeam irradiation. Proc Natl Acad Sci USA 44:553–559
Gavis ER, Singer RH, Huttelmaier S (2007) Localized translation through messenger RNA localization. In: Hershey JWB, Mathews MB, Sonenberg N (eds) Translational control in biology and medicine. Cold Spring Harbor Press, New York, pp 687–717
Groisman I, Huang YS, Mendez R et al (2000) CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103:435–447
Hartman H, Puma JP, Gurney T (1974) Evidence for the association of RNA with the ciliary basal bodies of Tetrahymena. J Cell Sci 16:241–259
Hiedemann SR, Sander G, Kirschner MW (1977) Evidence for a functional role of RNA in centrioles. Cell 10:337–350
Johnson KA, Rosenbaum JL (1991) Basal bodies and DNA. Trends Cell Biol 1:145–149
Kingsley EP, Chan XY, Duan Y, Lambert JD (2007) Widespread RNA segregation in a spiralian embryo. Evol Dev 9:527–539
Laane MM, Haugli FB (1974) Division centers in mitotic nuclei of Physarum polycephalum plasmodia. Nor J Bot 21:309–318
Lambert JD, Nagy LM (2002) Asymmetric inheritance of centrosomally-localized mRNAs during embryonic cleavages. Nature 420:682686
Lavdowsky M (1894) Formation of the chromatic and achromatic substances in animal and plant cells (in German). Merkel Bonnet’s Anat Hefte 4:13
Lécuyer E, Yoshida H, Parsatharathy N et al (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131:174–187
Loncarek J, Sluder G, Khodjakov A (2007) Centriole biogenesis: a tale of two pathways. Nat Cell Biol 9:736–738
Love R (1965) Differences in the internal structure of nucleoli in diploid and non-diploid transformed or neoplastic cells in vitro. Expt’ Cell Res 40:188–192
Love R (1966) Anisonucleolinosis in mammalian cell cultures. Natl Cancer Inst Monogr 23:167–180
Love R, Bharadwaj TP (1959) Two types of ribonucleoprotein in the nucleolus of mammalian cells. Nature 183:1453–1454
Love R, Liles RH (1959) Differentiation of nucleoproteins by inactivation of protein-bound amino groups and staining with toluidine blue and ammonium molybdate. J Histochem Cytochem 7:164–181
Love R, Soriano RZ (1971) Correlation of nucleolini with fine structural constituents of cultured normal and neoplastic cells. Cancer Res 31:1030–1037
Love R, Walsh RJ (1968) Nucleolinar morphology in normal diploid, neoplastic, and aneuploid cells in vitro. Cancer Res 30:990–997
Love R, Wildy P (1963) Cytochemical studies of the nucleoproteins of HeLa cells infected with herpes virus. J Cell Biol 17:237–254
Margulis LM, Chapman M, Dolan MF (2007) Semes for the analysis of evolution: deDuve’s peroxisomes and Myer's hydrogenases in the sulphurous Proterozoic eon. Nat Rev Gen 8:1–2
Marshall WF, Rosenbaum JL (2000) Are there nucleic acids in the centrosome? Curr Top Dev Biol 49:187–205
Mazia D (1955) The organization of the mitotic apparatus. Symp Soc Exp Biol 9:335–357
Montgomery TH (1898) Comparative cytological studies, with especial regard to the morphology of the nucleolus. J Morph 15:265–583
Oliver JM, Osbone WR, Pfeiffer JR et al (1981) Purine nucleoside phosphorylase is associated with centrioles and basal bodies. J Cell Biol 91:837–847
Pepper DA, Brinkley BR (1980) Tubulin nucleation and assembly in mitotic cells: evidence for nucleic acids in kinetochores and centrosomes. Cell Motil 1:1–15
Peterson SP, Berns MW (1978) Evidence for centriolar region RNA functioning in spindle formation in dividing PtK2 cells. J Cell Sci 34:289–301
Pettijohn DE, Hecht R (1974) RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol 38:31–41
Rieder CL (1979) Ribonucleoprotein staining of centrioles and kinetochores in newt lung cell spindles. J Cell Biol 80:1–9
Rustad RC (1959) An interference microscopical and cytochemical analysis of local mass changes in the mitotic apparatus during mitosis. Exp Cell Res 16:575–583
Shimamura T, Ota T (1956) Cytochemical studies on the mitotic spindle and the phragmoplast of plant cells. Exp Cell Res 11:346–361
Snyder JA (1980) Evidence for a ribonucleoprotein complex as a template for microtubule initiation in vivo. Cell Biol Int Rep 4:859–868
Stich H (1954) Substances and striations in the spindle of Cyclops strenuus; mechanism of mitosis. Chromosoma 6:199–236
Tsai M, Wang S, Heidinger JM et al (2006) A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311:1887–1893
Ugrinova I, Monier K, Ivaldi C et al (2007) Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8:66
Woodson SA (2008) RNA folding and ribosome assembly. Curr Opin Chem Biol 12:667–673
Wu X, Palazzo RE (1999) Differential regulation of maternal vs. paternal centrosomes. Proc Nat'l Acad Sci USA 96:1397–1402
Wulf E, Bautz FA, Faulstich H, Wieland T (1980) Distribution of fluorescent α-amanitin (FAMA) during mitosis in cultured rat kangaroo (PtK1) cells. Exp Cell Res 130:415–420
Zackroff RV, Rosenfeld AC, Weisenberg RC (1976) Effects of RNase and RNA on in vitro aster assembly. J Supramolec Struc 5:577–589
Zimmerman A (1960) Physico-chemical analysis of the isolated mitotic apparatus. Exp Cell Res 20:529–547
Acknowledgments
This work was supported by grants from the NIH (GM088503) and NSF (MCB0843092) to MCA. The author thanks Mary Anne Alliegro for reading and commenting on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: James Wakefield and Herbert Macgregor
Rights and permissions
About this article
Cite this article
Alliegro, M.C. The centrosome and spindle as a ribonucleoprotein complex. Chromosome Res 19, 367–376 (2011). https://doi.org/10.1007/s10577-011-9186-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9186-7