Abstract
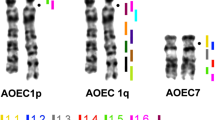
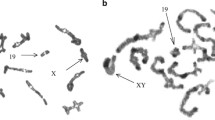
Monobrachial homology resulting from Robertsonian (Rb) fusions is thought to contribute to chromosomal speciation through underdominance. Given the karyotypic diversity characterizing wild house mouse populations [Mus musculus domesticus, (MMU)], variation that results almost exclusively from Rb fusions (diploid numbers range from 22 to 40) and possibly whole arm reciprocal translocations (WARTs), this organism represents an excellent model for testing hypotheses of chromosomal evolution. Previous studies of chromosome size and recombination rates have failed to explain the bias for certain chromosomes to be involved more frequently than others in these rearrangements. Here, we show that the pericentromeric region of one such chromosome, MMU19, which is infrequently encountered as a fusion partner in wild populations, is significantly enriched for housekeeping genes when compared to other chromosomes in the genome. These data suggest that there is selection against breakpoints in the pericentromeric region and provide new insights into factors that constrain chromosomal reorganizations in house mice. Given the anticipated increase in vertebrate whole genome sequences, the examination of gene content and expression profiles of the pericentromeric regions of other mammalian lineages characterized by Rb fusions (i.e., other rodents, bats, and bovids, among others) is both achievable and crucial to developing broadly applicable models of chromosome evolution.


Similar content being viewed by others
Abbreviations
- MMU:
-
Mus musculus domesticus
- Rb:
-
Robertsonian
- HSKP:
-
Housekeeping
- DSBs:
-
Double strand breaks
- PEV:
-
Position effect variegation
References
Baker RJ, Bickham JW (1986) Speciation by monobranchial centric fusions. Proc Natl Acad Sci USA 83:8245–8248
Bourque G, Pevzner PA, Tesler G (2004) Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res 14:507–516
Britton-Davidian J, Nadeau JH, Croset H, Thaler L (1989) Genetic differentiation and origin of Robertsonian populations of the house mouse (Mus musculus domesticus). Genet Res 53:29–44
Britton-Davidian J, Catalan J, Ramalhinho MD et al (2000) Rapid chromosomal evolution in island mice. Nature 403:158
Britton-Davidian J, Catalan J, Ramalhinho MDG et al (2005) Chromosomal phylogeny of Robertsonian races of the house mouse on the island of Madeira: testing between alternative mutational processes. Genet Res Camb 86:171–183
Butte AJ, Dzau VJ, Glueck SB (2001) Further defining housekeeping, or “maintenance”, genes. Focus on “a compendium of gene expression in normal human tissues”. Physiol Genomics 7:95–96
Cattanach BM (1974) Position effect variegation in the mouse. Genet Res 23:291–306
Cattanach BM (1975) Control of chromosome inactivation. Annu Rev Genet 9:1–18
Dobie KW, Leet M, Fantest JA et al (1996) Variegated transgene expression in mouse mammary gland is determined by the transgene integration locus. Proc Natl Acad Sci USA 93:6659–6664
Eisenberg E, Levanon EY (2003) Human housekeeping genes are compact. Trends Genet 19:362–365
Evans EP, Lyon MF, Daglish M (1967) A mouse translocation giving a metacentric marker chromosome. Cytogenet Cell Genet 6:105–119
Freilich S, Massingham T, Bhattacharyya S et al (2005) Relationship between the tissue-specificity of mouse gene expression and the evolutionary origin and function of the proteins. Genome Biol 6:R56
Garagna S, Broccoli D, Redi CA, Searle JB, Cooke HJ, Cappana E (1995) Robertsonian metacentrics of the house mouse lose telomeric sequences but retain some minor satellite sequences DNA in the pericentromeric area. Chromosoma 103:685–692
Garagna S, Marziliano N, Zuccotti M, Searle JB, Capanna E, Redi CA (2001) Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc Natl Acad Sci U S A 98:171–175
Garagna S, Zuccotti M, Capanna E, Redi CA (2002) High resolution organization of mouse telomeric and pericentromeric DNA. Cytogenet Genome Res 96:125–129
Gazave E, Catalan J, Ramalhinho MD et al (2003) The non-random occurrence of Robertsonian fusion in the house mouse. Genet Res 81:33–42
Kalitsis P, Griffiths B, Choo KH (2006) Mouse telomeric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc Natl Acad Sci U S A 103:8786–8791
King M (1993) Species evolution: the role of chromosome change. Cambridge University Press
Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA (2009) Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res 19:770–777
Lemaitre C, Zaghloul L, Sagot MF et al (2009) Analysis of fine-scale mammalian evolutionary breakpoints provides new insight into their relation to genome organisation. BMC Genomics 10:335
Nachman MW, Searle JB (1995) Why is the house mouse karyotype so variable? Trends Ecol Evol 10:397–402
Muller HJ (1930) Types of visible variations induced by X-rays in Drosophila. J Genet 22:299–334
Opsahl ML, McClenaghan M, Springbett A et al (2002) Multiple effects of genetic background on variegated transgene expression in mice. Genetics 160:1107–1112
Pedram M, Sprung CN, Gao Q, Lo AWI, Reynolds GE, Murnane JP (2006) Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol 26:1865–1878
Pialek J, Hauffe HC, Searle JB (2005) Chromosomal variation in the house mouse. Biol J Linn Soc 84:535–563
Qumsiyeh MB (1994) Evolution of number and morphology of mammalian chromosomes. J Hered 85:455–465
Redi CA, Capanna E (1988) Robertsonian heterozygotes in the house mouse and the fate of their germ cells. In: Liss AR (ed) The cytogenetics of mammalian autosomal rearrangements. pp 315–359
Rieseberg LH (2001) Chromosomal rearrangements and speciation. Trends Ecol Evol 16:351–358
Ruiz-Herrera A, Castresana J, Robinson TJ (2006) Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol 7:R115
Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E (2008) Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet Genome Res 122:219–228
Singer GAC, Lloyd AT, Huminiecki LB, Wolfe KH (2005) Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol Biol Evol 22:767–775
Slijepcevic P (1998) Telomeres and mechanisms of Robertsonian fusion. Chromosoma 107:136–140
Su AI, Wiltshire T, Batalov S et al (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 101:6062–6067
Vinogradov AE, Anatskaya OV (2007) Organismal complexity, cell differentiation and gene expression: human over mouse. Nucleic Acids Res 35:6350–6356
Wang Y, Rekaya R (2009) Comprehensive analysis of gene expression evolution between humans and mice. Evol Bioinform Online 5:81–90
White MJD (1978) Modes of speciation. Freeman, San Francisco
White BJ, Tjio JH (1968) A mouse translocation with 38 and 39 chromosomes but normal NF. Hereditas 58:284
Williams EJB, Hurst LD (2002) Clustering of tissue-specific genes underlies much of the similarity in rates of protein evolution of linked genes. J Mol Evol 54:511–518
Wu C, Orozco C, Boyer J et al (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10:R130
Zhao S, Shetty J, Hou L et al (2004) Human, mouse, and rat genome large-scale rearrangements: stability versus speciation. Genome Res 14:1851–1860
Zhimulev IF (1988) Polytene chromosomes, heterochromatin, and position effect variegation. Adv Genet 37:1–555
Zhu J, He F, Song S, Wang J, Yu J (2008) How many human genes can be defined as housekeeping with current expression data? BMC Genomics 9:172
Acknowledgements
Financial support from Ministerio de Ciencia y Tecnologia and the Universitat Autònoma de Barcelona (Ph.D. fellowship to M.F.) are gratefully acknowledged. T.J.R. is funded by a grant from the South African National Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert Macgregor.
Rights and permissions
About this article
Cite this article
Ruiz-Herrera, A., Farré, M., Ponsà, M. et al. Selection against Robertsonian fusions involving housekeeping genes in the house mouse: integrating data from gene expression arrays and chromosome evolution. Chromosome Res 18, 801–808 (2010). https://doi.org/10.1007/s10577-010-9153-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-010-9153-8