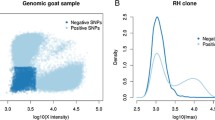
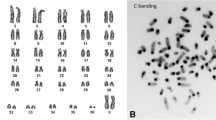
Abstract
A comprehensive physical map was generated for Ovis aries chromosome X (OARX) based on a cytogenomics approach. DNA probes were prepared from bacterial artificial chromosome (BAC) clones from the CHORI-243 sheep library and were assigned to G-banded metaphase spreads via fluorescence in-situ hybridization (FISH). A total of 22 BACs gave a single hybridization signal to the X chromosome and were assigned out of 32 tested. The positioned BACs contained 16 genes and a microsatellite marker which represent new cytogenetically mapped loci in the sheep genome. The gene and microsatellite loci serve to anchor between the existing radiation hybrid (RH) and virtual sheep genome (VSG) maps to the cytogenetic OARX map, whilst the BACs themselves also serve as anchors between the VSG and the cytogenetic maps. An additional 17 links between the RH and cytogenetic maps are provided by BAC end sequence (BES) derived markers that have also been positioned on the RH map. Comparison of the map orders for the cytogenetic, RH, and virtual maps reveals that the orders for the cytogenetic and RH maps are most similar, with only one locus, represented by BAC CH243-330E18, mapping to relatively different positions. Several discrepancies, including an inverted segment are found when comparing both the cytogenetic and RH maps with the virtual map. These discrepancies highlight the value of using physical mapping methods to inform the process of future in silico map construction. A detailed comparative analysis of sheep, human, and cattle mapping data allowed the construction of a comparative map that confirms and expands the knowledge about evolutionary conservation and break points between the X chromosomes of the three mammalian species.




Similar content being viewed by others
Abbreviations
- BAC:
-
bacterial artificial chromosome
- BES:
-
BAC end sequence
- BTA:
-
Bos taurus
- CATS:
-
comparative anchor tagged sequences
- FISH:
-
fluorescence in-situ hybridization
- FITC:
-
fluorescein isothiocyanate
- HSA:
-
Homo sapiens
- ISCNDB:
-
International System for Chromosome Nomenclature of Domestic Bovids
- OAR:
-
Ovis aries
- PI:
-
propidium iodide
- RH:
-
radiation hybrid
- VSG:
-
virtual sheep genome
References
Cockett NE (2006) The sheep genome. Genome Dyn 2:79–85
Cribiu EP, Di Berardino D, Di Meo GP et al (2001) International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet Cell Genet 92:283–299
Dalrymple BP, Kirkness EF, Nefedov M, International Sheep Genomics Consortium et al (2007) Using comparative genomics to reorder the human genome sequence into a virtual sheep genome. Genome Biol 8:R152
Di Meo GP, Perucatti A, Incarnato D et al (2002) Comparative mapping of twenty-eight bovine loci in sheep (Ovis aries, 2n = 54) and river buffalo (Bubalus bubalis, 2n = 50) by FISH. Cytogenet Genome Res 98:262–264
Di Meo GP, Perucatti A, Gautier M et al (2003) Chromosome localization of the 31 type I Texas bovine markers in sheep and goat chromosomes by comparative FISH-mapping and R-banding. Anim Genet 34:294–296
Di Meo GP, Perucatti A, Floriot S et al (2005) Chromosome evolution and improved cytogenetic maps of the Y chromosome in cattle, zebu, river buffalo, sheep and goat. Chromosome Res 13:349–355
Di Meo GP, Perucatti A, Floriot S et al (2007) An advanced sheep (Ovis aries, 2n = 54) cytogenetic map and assignment of 88 new autosomal loci by fluorescence in situ hybridization and R-banding. Anim Genet 38:233–240
Dobigny G, Yang F (2008) Foreword: Comparative cytogenetics in the genomics era: cytogenomics comes of age. Chromosome Res 16:1–4
Fortna A, Kim Y, MacLaren E et al (2004) Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol 2:E207
Goldammer T, Brunner RM, Weikard R, Kuehn C, Wimmers K (2007) Generation of an improved cytogenetic and comparative map of Bos taurus chromosome BTA27. Chromosome Res 15:203–213
Goldammer T, Brunner RM, Rebl A et al. (2009) A high-resolution radiation hybrid map of sheep chromosome X and comparison with human and cattle. Cytogenet Genome Res 124: (DOI:10.1159/000207520, in press).
Hecht J, Kuhl H, Haas SA et al (2006) Gene identification and analysis of transcripts differentially regulated in fracture healing by EST sequencing in the domestic sheep. BMC Genomics 7:172
Iannuzzi L, Di Meo GP, Perucatti A et al (2000) Comparative FISH-mapping of bovid X chromosomes reveals homologies and divercences between the subfamilies Bovinae and Caprinae. Cytogenet Cell Genet 89:171–176
Lyons LA, Laughlin TF, Copeland NG et al (1997) Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nature Genet 15:47–56
Maddox JF, Davies KP, Crawford AM et al (2001) An enhanced linkage map of the sheep genome comprising more than 1000 loci. Genome Res 11:1275–1289
Mandon-Pépin B, Oustry-Vaiman A, Vigier B, Piumi F, Cribiu E, Cotinot C (2003) Expression profiles and chromosomal localization of genes controlling meiosis and follicular development in the sheep ovary. Biol Reprod 68:985–995
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971
Tetens J, Goldammer T, Maddox JF, Cockett NE, Leeb T, Drögemüller C (2007) A radiation hybrid map of sheep chromosome 23 based on ovine BAC-end sequences. Anim Genet 38:132–140
Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S (2002) Guidelines for human gene nomenclature. Genomics 79:464–470
Watanabe H, Fujiyama A, Hattori M et al (2004) DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429:382–388
Wu CH, Nomura K, Goldammer T, Hadfield T, Womack JE, Cockett NE (2007) An ovine whole-genome radiation hybrid panel used to construct an RH map of ovine chromosome 9. Anim Genet 38:534–536
Wu CH, Hadfield T, Dalrymple BP et al (2009) Using the ovine radiation hybrid map to enhance comparisons of the ovine, bovine and human genomes. Plant & Animal Genomes XVII Conference, San Diego, CA, USA, p 496
Yunis JJ, Prakash O (1982) The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530
Acknowledgements
We gratefully thank Brigitte Schöpl, Ingrid Hennings, and Janine Vogt for excellent technical assistance in the mapping procedure. We thank Roland Riebe from FLI-Insel Riems for providing the embryonic sheep fibroblast cell line (SE-R). DFG-Grant GO-896/6-3 from the German Research Foundation supported the comparative mapping study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert Macgregor.
Accession numbers: Sequence data from this article have been deposited with the GenBank Data Library under Accession Nos. FJ853178–FJ853188 and FJ868495–FJ868499.
Rights and permissions
About this article
Cite this article
Goldammer, T., Brunner, R.M., Rebl, A. et al. Cytogenetic anchoring of radiation hybrid and virtual maps of sheep chromosome X and comparison of X chromosomes in sheep, cattle, and human. Chromosome Res 17, 497–506 (2009). https://doi.org/10.1007/s10577-009-9047-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-009-9047-9