Abstract
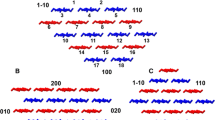
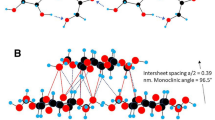
The effect of surface hydrophobicity and side-chain variation on xyloglucan adsorption onto cellulose microfibrils (CMF) is investigated via molecular dynamics simulations. A molecular model of CMF with (100), (010), (1–10), (110) and (200) crystal faces was built. We considered xylogluco-oligosaccharides (XGO) with three repeating units, namely (XXXG)3, (XXLG)3, and (XXFG)3 (where each (1,4)-β-d-glucosyl residue in the backbone is given a one-letter code according to its substituents: G = β-d-Glc; X = α-d-Xyl-(1,6)-β-d-Glc; L = β-d-Gal-(1,2)-α-d-Xyl-(1,6)-β-d-Glc; F = α-l-Fuc-(1,2)-β-d-Gal-(1,2)-α-d-Xyl-(1,6)-β-d-Glc). Our work shows that (XXXG)3 binds more favorably to the CMF (100) and (200) hydrophobic surfaces than to the (110), (010) and (1–10) hydrophilic surfaces. The origin of this behavior is attributed to the topography of hydrophobic CMF surface, which stabilizes (XXXG)3 in flat conformation. In contrast, on the rough hydrophilic CMF surface (XXXG)3 adopts a less favorable random-coil conformation to facilitate more hydrogen bonds with the surface. Extending the xyloglucan side chains from (XXXG)3 to (XXLG)3 hinders their stacking on the CMF hydrophobic surface. For (XXFG)3, the interaction with the hydrophobic surface is as strong as (XXXG)3. All three XGOs have similar binding to the hydrophilic surface. Steered molecular dynamics simulation was performed on an adhesive model where (XXXG)3 was sandwiched between two CMF hydrophobic surfaces. Our analysis suggests that this sandwich structure might help provide mechanical strength for plant cell walls. Our study relates to a recently revised model of primary cell walls in which extensibility is largely determined by xyloglucan located in limited regions of tight contact between CMFs.










Similar content being viewed by others
References
Bacic A, Harris P, Stone B, Preiss J (1988) Structure and function of plant cell walls. Biochem Plant 14:297–371
Besombes S, Mazeau K (2005) The cellulose/lignin assembly assessed by molecular modeling. Part 1: adsorption of a threo guaiacyl β-O-4 dimer onto a Iβ cellulose whisker. Plant Physiol Biochem 43:299–308
Brooks B, Brooks C, Mackerell AD, MacKerell A et al (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614
Brown Jr RM (1990) Microbial cellulose modified during synthesis. U.S. Patent No. 4,942,128
Brumer H, Zhou Q, Baumann M et al (2004) Activation of crystalline cellulose surfaces through the chemoenzymatic modification of xyloglucan. J Am Chem Soc 126:5715–5721
Burgert I (2006) Exploring the micromechanical design of plant cell walls. Am J Bot 93:1391–1401
Carpita N (1985) Tensile strength of cell walls of living cells. J Plant Physiol 79:485–488
Cavalier D, Lerouxel O (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell Online 20:1519–1537
Cosgrove D (1993) Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol 124:1–23
Cosgrove D (2005) Growth of the plant cell wall. Nat Rev Mol Cell Bio 6:850–861
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an Nlog (N) method for Ewald sums in large systems. J Chem Phys 98:10089
de Lima D, Buckeridge M (2001) Interaction between cellulose and storage xyloglucans: the influence of the degree of galactosylation. Carbohydr Polym 46:157–163
Desveaux D, Faik A, Maclachlan G (1998) Fucosyltransferase and the biosynthesis of storage and structural xyloglucan in developing nasturtium fruits. Plant Physiol 118:885–894
Dick-Pérez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M (2011) Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50:989–1000
Dufresne A (2008) Polysaccharide nano crystal reinforced nanocomposites. Can J Chem 86:484–494
Durell SR, Brooks BR, Ben-Naim A (1994) Solvent-induced forces between two hydrophilic groups. J Phys Chem 98:2198–2202
Essmann U, Perera L, Berkowitz ML et al (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108:1195–1203
Fink H, Ahrenstedt L, Bodin A (2011) Bacterial cellulose modified with xyloglucan bearing the adhesion peptide RGD promotes endothelial cell adhesion and metabolism—a promising modification for vascular grafts. J Tissue Eng Regen Med 5:454–463
Fry S (1989) The structure and functions of xyloglucan. J Exp Bot 40:1–11
Guvench O, Hatcher ER, Venable RM et al (2009) CHARMM additive all-atom force field for acyclic polyalcohols, acyclic carbohydrates and inositol. J Chem Theory Comp 5:2353–2370
Guzman D, Roland J, Keer H et al (2008) Using steered molecular dynamics simulations and single-molecule force spectroscopy to guide the rational design of biomimetic modular polymeric materials. Polymer 49:3892–3901
Hanley S, Revol J, Godbout L et al (1997) Atomic force microscopy and transmission electron microscopy of cellulose from Micrasterias denticulata; evidence for a chiral helical microfbril twist. Cellulose 4:209–220
Hanus J, Mazeau K (2006) The xyloglucan–cellulose assembly at the atomic scale. Biopolymers 82:59–73
Hayashi T (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Biol 40:139–168
Hayashi T, Takeda T (1994) Effects of the degree of polymerization on the binding of xyloglucans to cellulose. Plant Cell Physiol 35:893–899
Hayashi T, Marsden M, Delmer D (1987) Pea xyloglucan and cellulose VI. Xyloglucan-cellulose interactions in vitro and in vivo. Plant Physiol 83:384–389
Helbert W, Nishiyama Y (1998) Molecular imaging of Halocynthia papillosa cellulose. J Struct Biol 124:42–50
Helbert W, Sugiyama J, Kimura S, Itoh T (1998) High-resolution electron microscopy on ultrathin sections of cellulose microfibrils generated by glomerulocytes in Polyzoa vesiculiphora. Protoplasma 203:84–90
Himmel M, Ruth M, Wyman C (1999) Cellulase for commodity products from cellulosic biomass. Curr Opin Biotechnol 10:358–364
Hisamatsu M, York W, Darvill A, Albersheim P (1992) Characterization of seven xyloglucan oligosaccharides containing from seventeen to twenty glycosyl residues. Carbohyd Res 227:45–71
Hubbe M, Rojas O, Lucia L, Sain M (2008) Cellulosic nanocomposites: a review. Bioresources 3:929–980
Izrailev S, Stepaniants S, Balsera M et al (1997) Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J 72:1568–1581
Jean B, Heux L, Dubreuil F, Chambat G, Cousin F (2009) Non-electrostatic building of biomimetic cellulose-xyloglucan multilayers. Langmuir 25:3920–3923
Jorgensen WL, Chandrasekhar JM, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kang Y, Liu YC, Wang Q, Shen JW, Wu T, Guan W (2009) On the spontaneous encapsulation of proteins in carbon nanotubes. Biomaterials 30:2807–2815
Kirschner K (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J Comput Chem 29:622–655
Krishnan R, Binkley J (1980) Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J Chem Phys 72:650
Levy S, York WS, Stuike-Pril R, Meyer B, Staehelin A (1991) Simulations of the static and dynamic molecular conformations of xyloglucan. The role of the fucosylated sidechain in surface-specific sidechain folding. Plant J 1:195–215
Levy S, Maclachlan G, Staehelin A (1997) Xyloglucan sidechains modulate binding to cellulose during in vitro binding assays as predicted by conformational dynamics simulations. Plant J 11:373–386
Lima D, Loh W, Buckeridge M (2004) Xyloglucan–cellulose interaction depends on the sidechains and molecular weight of xyloglucan. Plant Physiol Biochem 42:389–394
Lopez M, Bizot H, Chambat G (2010) Enthalpic studies of xyloglucan-cellulose interactions. Biomacromolecules 11:1417–1428
Madson M, Dunand C, Li X (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell Online 7:1662–1670
Mazeau K (2011) On the external morphology of native cellulose microfibrils. Carbohydr Polym 84:524–532
Mazeau K, Vergelati C (2002) Atomistic modeling of the adsorption of benzophenone onto cellulosic surfaces. Langmuir 18:1919–1927
Mazeau K, Wyszomirski M (2012) Modelling of Congo red adsorption on the hydrophobic surface of cellulose using molecular dynamics. Cellulose 19:1495–1506
McNeil M, Darvill A (1984) Structure and function of the primary cell walls of plants. Annu Rev Biochem 53:625–663
Mishra A, Malhotra AV (2009) Tamarind xyloglucan: a polysaccharide with versatile application potential. J Mater Chem 19:8528–8536
Moon R, Martini A, Nairn J (2011) Cellulose nanomaterials review: structure, properties and nanocomposites. Chem Soc Rev 40:3941–3994
Morfill J, Neumann J, Blank K et al (2008) Force-based analysis of multidimensional energy landscapes: application of dynamic force spectroscopy and steered molecular dynamics simulations to an antibody fragment–peptide complex. J Mol Biol 381:1253–1266
Nelson M, Humphrey W (1996) NAMD: a parallel, object-oriented molecular dynamics program. J High Perform Comput Appl 10:251–268
Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124:9074–9082
Park Y, Cosgrove D (2012) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158:1933–1943
Pauly M, Andersen L, Kauppinen S (1999) A xyloglucan-specific endo-β-1, 4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9:93–100
Peña M, Ryden P, Madson M (2004) The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol 134:443–451
Pérez S, Mazeau K (2005) Conformations, structures, and morphologies of celluloses. In: Dumitriu S (ed) Polysaccharides: structural diversity and functional versatility, 2nd edn. Dekker, New York, pp 41–68
Ryden P, Sugimoto-Shirasu K (2003) Tensile properties of Arabidopsis cell walls depend on both a xyloglucan cross-linked microfibrillar network and rhamnogalacturonan II-borate complexes. Plant Physiol 132:1033–1040
Shen JW, Wu T, Wang Q, Pan HH (2008) Induced stepwise conformational change of human serum albumin on carbon nanotube surfaces. Biomaterials 29:513–532
Sugiyama J, Vuong R, Chanzy H (1991) Electron diffraction study on the two crystalline phases occurring in native cellulose from an algal cell wall. Macromolecules 24:4168–4175
Sun Y, Cheng J (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83:1–11
Van Daele Y, Revol J, Gaill F, Goffinet G (1992) Characterization and supramolecular architecture of the cellulose-protein fibrils in the tunic of the sea peach (Halocynthia papillosa, Ascidiacea, Urochordata). Biol Cell 76:87–96
Van Gunsteren W, Berendsen H (1977) Algorithms for macromolecular dynamics and constraint dynamics. Mol Phys 34:1311–1327
Vanzin G, Madson M (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99:3340–3345
Vincken JP, de Keizer A, de Keizer A et al (1995) Fractionation of xyloglucan fragments and their interaction with cellulose. Plant Physiol 108:1579–1585
Wegner T, Jones E (2009) A fundamental review of the relationships between nanotechnology and lignocellulosic biomass. Nanosci Technol Renew Biomater 1:1–41
Zhang Q, Brumer H, Ågren H, Tu Y (2011) The adsorption of xyloglucan on cellulose: effects of explicit water and side chain variation. Carbohydr Res 346:2595–2602
Zhao Y, Truhlar D (2006) A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interactions. J Chem Phys 125:194101
Zhao Y, Truhlar D (2007) Density functionals for noncovalent interaction energies of biological importance. J Chem Theory Comput 3:289–300
Acknowledgments
This work is supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC0001090. We acknowledge the Penn State University RCC Center for supercomputer time.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Z., Crespi, V.H., Kubicki, J.D. et al. Molecular dynamics simulation study of xyloglucan adsorption on cellulose surfaces: effects of surface hydrophobicity and side-chain variation. Cellulose 21, 1025–1039 (2014). https://doi.org/10.1007/s10570-013-0041-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-013-0041-1