Abstract
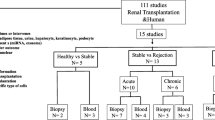
For end-stage renal diseases, kidney transplantation is the most efficient treatment. However, the unexpected rejection caused by inflammation usually leads to allograft failure. Thus, a systems-level characterization of inflammation factors can provide potentially diagnostic biomarkers for predicting renal allograft rejection. Serum of kidney transplant patients with different immune status were collected and classified as transplant patients with stable renal function (ST), impaired renal function with negative biopsy pathology (UNST), acute rejection (AR), and chronic rejection (CR). The expression profiles of 40 inflammatory proteins were measured by quantitative protein microarrays and reduced to a lower dimensional space by the partial least squares (PLS) model. The determined principal components (PCs) were then trained by the support vector machines (SVMs) algorithm for classifying different phenotypes of kidney transplantation. There were 30, 16, and 13 inflammation proteins that showed statistically significant differences between CR and ST, CR and AR, and CR and UNST patients. Further analysis revealed a protein-protein interaction (PPI) network among 33 inflammatory proteins and proposed a potential role of intracellular adhesion molecule-1 (ICAM-1) in CR. Based on the network analysis and protein expression information, two PCs were determined as the major contributors and trained by the PLS-SVMs method, with a promising accuracy of 77.5 % for classification of chronic rejection after kidney transplantation. For convenience, we also developed software packages of GPS-CKT (Classification phenotype of Kidney Transplantation Predictor) for classifying phenotypes. By confirming a strong correlation between inflammation and kidney transplantation, our results suggested that the network biomarker but not single factors can potentially classify different phenotypes in kidney transplantation.





Similar content being viewed by others
Abbreviations
- AR:
-
Acute rejection
- BLC:
-
B lymphocyte chemoattractant
- CR:
-
Chronic rejection
- G-CSF:
-
Granulocyte colony-stimulating factor
- GM-CSF:
-
Granulocyte/macrophage colony-stimulating factor
- UNST:
-
Impaired renal function with negative biopsy pathology
- ICAM-1:
-
Intracellular adhesion molecule-1
- LOO:
-
Leave-one-out
- PLS:
-
Partial least squares
- PCs:
-
Principal components
- PPI:
-
Protein-protein interaction
- M-CSF:
-
Macrophage colony-stimulating factor-1
- TIMP-2:
-
Metalloproteinase inhibitor 2
- MCP-1:
-
Monocyte chemoattractant protein-1
- ROC:
-
Receiver operation characteristic
- ST:
-
Stable renal function
- SVMs:
-
Support vector machines
References
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 2003;4:2.
Beckingham IJ, Nicholson ML, Bell PR. Analysis of factors associated with complications following renal transplant needle core biopsy. Br J Urol. 1994;73:13–5.
Chang CC, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(27):1–27.
Chen Y, Jiang T, Jiang R. Uncover disease genes by maximizing information flow in the phenome-interactome network. Bioinformatics. 2011;27:i167–76.
Chen H, Song Z, Qian M, Bai C, Wang X. Selection of disease-specific biomarkers by integrating inflammatory mediators with clinical informatics in AECOPD patients: a preliminary study. J Cell Mol Med. 2012;16:1286–97.
Gallagher MP, Kelly PJ, Jardine M, Perkovic V, Cass A, Craig JC, et al. Long-term cancer risk of immunosuppressive regimens after kidney transplantation. J Am Soc Nephrol. 2010;21:852–8.
Hartono C, Muthukumar T, Suthanthiran M. Noninvasive diagnosis of acute rejection of renal allografts. Curr Opin Organ Transplant. 2010;15:35–41.
Heidt S, San Segundo D, Shankar S, Mittal S, Muthusamy AS, Friend PJ, et al. Peripheral blood sampling for the detection of allograft rejection: biomarker identification and validation. Transplantation. 2011;92:1–9.
Hill PA, Lan HY, Nikolic-Paterson DJ, Atkins RC. ICAM-1 directs migration and localization of interstitial leukocytes in experimental glomerulonephritis. Kidney Int. 1994;45:32–42.
Jin G, Zhou X, Wang H, Zhao H, Cui K, Zhang XS, et al. The knowledge-integrated network biomarkers discovery for major adverse cardiac events. J Proteome Res. 2008;7:4013–21.
Krensky AM, Ahn YT. Mechanisms of disease: regulation of RANTES (CCL5) in renal disease. Nat Clin Pract Nephrol. 2007;3:164–70.
Musial K, Zwolinska D. TIMP-2. Cell Stress Chaperones. 2011;16:97–103.
Nguyen DV, Rocke DM. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics. 2002;18:39–50.
Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012:40, D71-5.
Rush DN, Henry SF, Jeffery JR, Schroeder TJ, Gough J. Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation. 1994;57:208–11.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2.
von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, et al. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–62.
Wilczek HE. Percutaneous needle biopsy of the renal allograft. A clinical safety evaluation of 1129 biopsies. Transplantation. 1990;50:790–7.
Wu D, Zhu D, Xu M, Rong R, Tang Q, Wang X, et al. Analysis of transcriptional factors and regulation networks in patients with acute renal allograft rejection. J Proteome Res. 2011;10:175–81.
Wu D, Rice CM, Wang X. Cancer bioinformatics: a new approach to systems clinical medicine. BMC Bioinforma. 2012a;13:71.
Wu D, Qi G, Wang X, Xu M, Rong R, Zhu T. Hematopoietic stem cell transplantation induces immunologic tolerance in renal transplant patients via modulation of inflammatory and repair processes. J Transl Med. 2012b;10:182.
Wu D, Liu X, Liu C, Liu Z, Xu M, Rong R, et al. Network analysis reveals roles of inflammatory factors in different phenotypes of kidney transplant patients. J Theor Biol. 2014;362:62–8.
Zhang SH, Wu C, Li X, Chen X, Jiang W, Gong BS, et al. From phenotype to gene: detecting disease-specific gene functional modules via a text-based human disease phenotype network construction. FEBS Lett. 2010;584:3635–43.
Acknowledgments
This work was sponsored by the National Natural Science Foundation of China (81500569) to D.Z.; National Natural Science Foundation of China (81100534) and Shanghai Rising-Star Program (13QA1400800) to D.W.; grants from the National Basic Research Program (973 project) (2012CB910101 and 2013CB933903), and Natural Science Foundation of China (31171263, 81272578) to Y.X. The authors have no commercial or other associations that might pose a conflict of interest in connection with the submitted material.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All samples were obtained with informed consent and ethics approval by the ethics board of Zhongshan Hospital, Fudan University. The study has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans.
Additional information
Dong Zhu and Zexian Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhu, D., Liu, Z., Pan, Z. et al. A new method for classifying different phenotypes of kidney transplantation. Cell Biol Toxicol 32, 323–332 (2016). https://doi.org/10.1007/s10565-016-9337-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-016-9337-x