Abstract
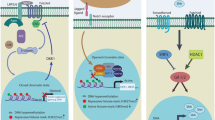
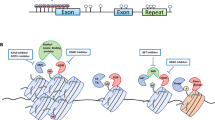
Cancer remains one of the most challenging diseases despite significant advances of early diagnosis and therapeutic treatments. Cancerous tumors are composed of various cell types including cancer stem cells capable of self-renewal, proliferation, differentiation, and invasion of distal tumor sites. Most notably, these cells can enter a dormant cellular state that is resistant to conventional therapies. Thereby, cancer stem cells have the intrinsic potential for tumor initiation, tumor growth, metastasis, and tumor relapse after therapy. Both genetic and epigenetic alterations are attributed to the formation of multiple tumor types. This review is focused on how epigenetic dynamics involving DNA methylation and DNA oxidations are implicated in breast cancer and glioblastoma multiforme. The emergence and progression of these cancer types rely on cancer stem cells with the capacity to enter quiescence also known as a dormant cellular state, which dictates the distinct tumorigenic aggressiveness between breast cancer and glioblastomas.




Similar content being viewed by others
References
Afifi, A. M., Saad, A. M., Al-Husseini, M. J., Elmehrath, A. O., Northfelt, D. W., & Sonbol, M. B. (2019). Causes of death after breast cancer diagnosis: a US population-based analysis. Cancer. https://doi.org/10.1002/cncr.32648.
Tamimi, A. F., & Juweid, M. (2017). Epidemiology and outcome of glioblastoma. In S. De Vleeschouwer (Ed.), Glioblastoma Brisbane (AU). Chapter 8, 143–154
Lathia, J. D., Mack, S. C., Mulkearns-Hubert, E. E., Valentim, C. L., & Rich, J. N. (2015). Cancer stem cells in glioblastoma. Genes & Development, 29(12), 1203–1217. https://doi.org/10.1101/gad.261982.115.
Omuro, A., & DeAngelis, L. M. (2013). Glioblastoma and other malignant gliomas: a clinical review. JAMA, 310(17), 1842–1850. https://doi.org/10.1001/jama.2013.280319.
Kahlert, U. D., Mooney, S. M., Natsumeda, M., Steiger, H. J., & Maciaczyk, J. (2017). Targeting cancer stem-like cells in glioblastoma and colorectal cancer through metabolic pathways. International Journal of Cancer, 140(1), 10–22. https://doi.org/10.1002/ijc.30259.
Han, H. R., Park, S. A., Ahn, S., Jeun, S. S., & Ryu, C. H. (2019). Evaluation of combination treatment effect with TRAIL-secreting mesenchymal stem cells and compound C against glioblastoma. Anticancer Research, 39(12), 6635–6643. https://doi.org/10.21873/anticanres.13878.
Pantel, K., Alix-Panabieres, C., & Riethdorf, S. (2009). Cancer micrometastases. Nature Reviews. Clinical Oncology, 6(6), 339–351. https://doi.org/10.1038/nrclinonc.2009.44.
Patel, S. A., Ramkissoon, S. H., Bryan, M., Pliner, L. F., Dontu, G., Patel, P. S., et al. (2012). Delineation of breast cancer cell hierarchy identifies the subset responsible for dormancy. Scientific Reports, 2, 906–906. https://doi.org/10.1038/srep00906.
Chrun, E. S., Modolo, F., & Daniel, F. I. (2017). Histone modifications: a review about the presence of this epigenetic phenomenon in carcinogenesis. Pathology, Research and Practice, 213(11), 1329–1339. https://doi.org/10.1016/j.prp.2017.06.013.
Ben-Porath, I., Thomson, M. W., Carey, V. J., Ge, R., Bell, G. W., Regev, A., et al. (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature Genetics, 40(5), 499–507. https://doi.org/10.1038/ng.127.
Suva, M. L., Riggi, N., & Bernstein, B. E. (2013). Epigenetic reprogramming in cancer. Science, 339(6127), 1567–1570. https://doi.org/10.1126/science.1230184.
Loh, K. M., & Lim, B. (2011). A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell, 8(4), 363–369. https://doi.org/10.1016/j.stem.2011.03.013.
Thomson, M., Liu, S. J., Zou, L. N., Smith, Z., Meissner, A., & Ramanathan, S. (2011). Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell, 145(6), 875–889. https://doi.org/10.1016/j.cell.2011.05.017.
Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676. https://doi.org/10.1016/j.cell.2006.07.024.
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131(5), 861–872. https://doi.org/10.1016/j.cell.2007.11.019.
Iwafuchi-Doi, M., & Zaret, K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes & Development, 28(24), 2679–2692. https://doi.org/10.1101/gad.253443.114.
Zhang, W., Bado, I., Wang, H., Lo, H. C., & Zhang, X. H. (2019). Bone metastasis: find your niche and fit in. Trends Cancer, 5(2), 95–110. https://doi.org/10.1016/j.trecan.2018.12.004.
Rowland, B. D., Bernards, R., & Peeper, D. S. (2005). The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nature Cell Biology, 7(11), 1074–1082. https://doi.org/10.1038/ncb1314.
Utikal, J., Polo, J. M., Stadtfeld, M., Maherali, N., Kulalert, W., Walsh, R. M., et al. (2009). Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature, 460(7259), 1145–1148. https://doi.org/10.1038/nature08285.
Hanna, J., Saha, K., Pando, B., van Zon, J., Lengner, C. J., Creyghton, M. P., et al. (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature, 462(7273), 595–601. https://doi.org/10.1038/nature08592.
Hochedlinger, K., & Jaenisch, R. (2015). Induced pluripotency and epigenetic reprogramming. Cold Spring Harbor Perspectives in Biology, 7(12). https://doi.org/10.1101/cshperspect.a019448.
De Carvalho, D. D., You, J. S., & Jones, P. A. (2010). DNA methylation and cellular reprogramming. Trends in Cell Biology, 20(10), 609–617. https://doi.org/10.1016/j.tcb.2010.08.003.
Gao, Y., Chen, J., Li, K., Wu, T., Huang, B., Liu, W., et al. (2013). Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell, 12(4), 453–469. https://doi.org/10.1016/j.stem.2013.02.005.
Sardina, J. L., Collombet, S., Tian, T. V., Gomez, A., Di Stefano, B., Berenguer, C., et al. (2018). Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell, 23(5), 727–741.e729. https://doi.org/10.1016/j.stem.2018.08.016.
Du, Q., Wang, Z., & Schramm, V. L. (2016). Human DNMT1 transition state structure. Proceedings of the National Academy of Sciences of the United States of America, 113(11), 2916–2921. https://doi.org/10.1073/pnas.1522491113.
Okashita, N., Kumaki, Y., Ebi, K., Nishi, M., Okamoto, Y., Nakayama, M., et al. (2014). PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development, 141(2), 269–280. https://doi.org/10.1242/dev.099622.
Bostick, M., Kim, J. K., Esteve, P. O., Clark, A., Pradhan, S., & Jacobsen, S. E. (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science, 317(5845), 1760–1764. https://doi.org/10.1126/science.1147939.
Stathopoulou, A., Chhetri, J. B., Ambrose, J. C., Esteve, P. O., Ji, L., Erdjument-Bromage, H., et al. (2017). A novel requirement for DROSHA in maintenance of mammalian CG methylation. Nucleic Acids Research, 45(16), 9398–9412. https://doi.org/10.1093/nar/gkx695.
Grosser, C., Wagner, N., Grothaus, K., & Horsthemke, B. (2015). Altering TET dioxygenase levels within physiological range affects DNA methylation dynamics of HEK293 cells. Epigenetics, 10(9), 819–833. https://doi.org/10.1080/15592294.2015.1073879.
Spruijt, C. G., Gnerlich, F., Smits, A. H., Pfaffeneder, T., Jansen, P. W., Bauer, C., et al. (2013). Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell, 152(5), 1146–1159. https://doi.org/10.1016/j.cell.2013.02.004.
Liu, L., Mao, S. Q., Ray, C., Zhang, Y., Bell, F. T., Ng, S. F., et al. (2015). Differential regulation of genomic imprinting by TET proteins in embryonic stem cells. Stem Cell Research, 15(2), 435–443. https://doi.org/10.1016/j.scr.2015.08.010.
Skiles, W. M., Kester, A., Pryor, J. H., Westhusin, M. E., Golding, M. C., & Long, C. R. (2018). Oxygen-induced alterations in the expression of chromatin modifying enzymes and the transcriptional regulation of imprinted genes. Gene Expression Patterns, 28, 1–11. https://doi.org/10.1016/j.gep.2018.01.001.
Li, X. L., Xu, J. H., Nie, J. H., & Fan, S. J. (2012). Exogenous expression of UHRF1 promotes proliferation and metastasis of breast cancer cells. Oncology Reports, 28(1), 375–383. https://doi.org/10.3892/or.2012.1792.
Unoki, M., Brunet, J., & Mousli, M. (2009). Drug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochemical Pharmacology, 78(10), 1279–1288. https://doi.org/10.1016/j.bcp.2009.05.035.
Maenohara, S., Unoki, M., Toh, H., Ohishi, H., Sharif, J., Koseki, H., et al. (2017). Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genetics, 13(10), e1007042. https://doi.org/10.1371/journal.pgen.1007042.
Gordon, C. A., Hartono, S. R., & Chedin, F. (2013). Inactive DNMT3B splice variants modulate de novo DNA methylation. PLoS One, 8(7), e69486. https://doi.org/10.1371/journal.pone.0069486.
Noh, K. M., Wang, H., Kim, H. R., Wenderski, W., Fang, F., Li, C. H., et al. (2015). Engineering of a histone-recognition domain in Dnmt3a alters the epigenetic landscape and phenotypic features of mouse ESCs. Molecular Cell, 59(1), 89–103. https://doi.org/10.1016/j.molcel.2015.05.017.
Jeltsch, A., & Jurkowska, R. Z. (2016). Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Research, 44(18), 8556–8575. https://doi.org/10.1093/nar/gkw723.
Duvall-Noelle, N., Karwandyar, A., Richmond, A., & Raman, D. (2016). LASP-1: a nuclear hub for the UHRF1-DNMT1-G9a-Snail1 complex. Oncogene, 35(9), 1122–1133. https://doi.org/10.1038/onc.2015.166.
Xue, B., Zhao, J., Feng, P., Xing, J., Wu, H., & Li, Y. (2019). Epigenetic mechanism and target therapy of UHRF1 protein complex in malignancies. Oncotargets and Therapy, 12, 549–559. https://doi.org/10.2147/ott.S192234.
Snowden, A. W., Gregory, P. D., Case, C. C., & Pabo, C. O. (2002). Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Current Biology, 12(24), 2159–2166. https://doi.org/10.1016/s0960-9822(02)01391-x.
Ko, M., An, J., Pastor, W. A., Koralov, S. B., Rajewsky, K., & Rao, A. (2015). TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunological Reviews, 263(1), 6–21. https://doi.org/10.1111/imr.12239.
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324(5929), 930–935. https://doi.org/10.1126/science.1170116.
Hu, X., Zhang, L., Mao, S. Q., Li, Z., Chen, J., Zhang, R. R., et al. (2014). Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell, 14(4), 512–522. https://doi.org/10.1016/j.stem.2014.01.001.
Liu, X. S., Wu, H., Ji, X., Stelzer, Y., Wu, X., Czauderna, S., et al. (2016). Editing DNA methylation in the mammalian genome. Cell, 167(1), 233–247.e217. https://doi.org/10.1016/j.cell.2016.08.056.
Wu, M. J., Kim, M. R., Chen, Y. S., Yang, J. Y., & Chang, C. J. (2017). Retinoic acid directs breast cancer cell state changes through regulation of TET2-PKCzeta pathway. Oncogene, 36(22), 3193–3206. https://doi.org/10.1038/onc.2016.467.
Xu, Y., Liu, S. Y., Li, J., Zhang, L., Chen, D., Zhang, J. P., et al. (2018). Real-time sensing of TET2-mediated DNA demethylation in vitro by metal-organic framework-based oxygen sensor for mechanism analysis and stem-cell behavior prediction. Analytical Chemistry, 90(15), 9330–9337. https://doi.org/10.1021/acs.analchem.8b01941.
Rasmussen, K. D., & Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes & Development, 30(7), 733–750. https://doi.org/10.1101/gad.276568.115.
Wang, L., Zhou, Y., Xu, L., Xiao, R., Lu, X., Chen, L., et al. (2015). Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature, 523(7562), 621–625. https://doi.org/10.1038/nature14482.
Kellinger, M. W., Song, C. X., Chong, J., Lu, X. Y., He, C., & Wang, D. (2012). 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nature Structural & Molecular Biology, 19(8), 831–833. https://doi.org/10.1038/nsmb.2346.
Globisch, D., Munzel, M., Muller, M., Michalakis, S., Wagner, M., Koch, S., et al. (2010). Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One, 5(12), e15367. https://doi.org/10.1371/journal.pone.0015367.
Zhang, L. Y., Han, C. S., Li, P. L., & Zhang, X. C. (2016). 5-Hydroxymethylcytosine expression is associated with poor survival in cervical squamous cell carcinoma. Japanese Journal of Clinical Oncology, 46(5), 427–434. https://doi.org/10.1093/jjco/hyw002.
Zhang, P., Huang, B., Xu, X., & Sessa, W. C. (2013). Ten-eleven translocation (Tet) and thymine DNA glycosylase (TDG), components of the demethylation pathway, are direct targets of miRNA-29a. Biochemical and Biophysical Research Communications, 437(3), 368–373. https://doi.org/10.1016/j.bbrc.2013.06.082.
Putiri, E. L., Tiedemann, R. L., Thompson, J. J., Liu, C., Ho, T., Choi, J. H., et al. (2014). Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biology, 15(6), R81. https://doi.org/10.1186/gb-2014-15-6-r81.
Iyer, L. M., Abhiman, S., & Aravind, L. (2011). Natural history of eukaryotic DNA methylation systems. Progress in Molecular Biology and Translational Science, 101, 25–104. https://doi.org/10.1016/b978-0-12-387685-0.00002-0.
Iyer, L. M., Tahiliani, M., Rao, A., & Aravind, L. (2009). Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle, 8(11), 1698–1710. https://doi.org/10.4161/cc.8.11.8580.
Ko, M., An, J., Bandukwala, H. S., Chavez, L., Aijo, T., Pastor, W. A., et al. (2013). Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature, 497(7447), 122–126. https://doi.org/10.1038/nature12052.
Nguyen, A. V., Albers, C. G., & Holcombe, R. F. (2010). Differentiation of tubular and villous adenomas based on Wnt pathway-related gene expression profiles. International Journal of Molecular Medicine, 26(1), 121–125. https://doi.org/10.3892/ijmm_00000443.
Zhang, P., Rausch, C., Hastert, F. D., Boneva, B., Filatova, A., Patil, S. J., et al. (2017). Methyl-CpG binding domain protein 1 regulates localization and activity of Tet1 in a CXXC3 domain-dependent manner. Nucleic Acids Research, 45(12), 7118–7136. https://doi.org/10.1093/nar/gkx281.
Ludwig, A. K., Zhang, P., Hastert, F. D., Meyer, S., Rausch, C., Herce, H. D., et al. (2017). Binding of MBD proteins to DNA blocks Tet1 function thereby modulating transcriptional noise. Nucleic Acids Research, 45(5), 2438–2457. https://doi.org/10.1093/nar/gkw1197.
Rausch, C., Hastert, F. D., & Cardoso, M. C. (2019). DNA modification readers and writers and their interplay. Journal of Molecular Biology. https://doi.org/10.1016/j.jmb.2019.12.018.
Li, L., Chen, B. F., & Chan, W. Y. (2015). An epigenetic regulator: methyl-CpG-binding domain protein 1 (MBD1). International Journal of Molecular Sciences, 16(3), 5125–5140. https://doi.org/10.3390/ijms16035125.
Wood, K. H., & Zhou, Z. (2016). Emerging molecular and biological functions of MBD2, a reader of DNA methylation. Frontiers in Genetics, 7, 93. https://doi.org/10.3389/fgene.2016.00093.
Takai, H., Masuda, K., Sato, T., Sakaguchi, Y., Suzuki, T., Suzuki, T., et al. (2014). 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Reports, 9(1), 48–60. https://doi.org/10.1016/j.celrep.2014.08.071.
Forloni, M., Gupta, R., Nagarajan, A., Sun, L. S., Dong, Y., Pirazzoli, V., et al. (2016). Oncogenic EGFR represses the TET1 DNA demethylase to induce silencing of tumor suppressors in cancer cells. Cell Reports, 16(2), 457–471. https://doi.org/10.1016/j.celrep.2016.05.087.
Good, C. R., Madzo, J., Patel, B., Maegawa, S., Engel, N., Jelinek, J., et al. (2017). A novel isoform of TET1 that lacks a CXXC domain is overexpressed in cancer. Nucleic Acids Research, 45(14), 8269–8281. https://doi.org/10.1093/nar/gkx435.
Inoue-Choi, M., Nelson, H. H., Robien, K., Arning, E., Bottiglieri, T., Koh, W. P., et al. (2013). Plasma S-adenosylmethionine, DNMT polymorphisms, and peripheral blood LINE-1 methylation among healthy Chinese adults in Singapore. BMC Cancer, 13, 389. https://doi.org/10.1186/1471-2407-13-389.
Kohli, R. M., & Zhang, Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature, 502(7472), 472–479. https://doi.org/10.1038/nature12750.
Cavuoto, P., & Fenech, M. F. (2012). A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treatment Reviews, 38(6), 726–736. https://doi.org/10.1016/j.ctrv.2012.01.004.
Soda, K. (2018). Polyamine metabolism and gene methylation in conjunction with one-carbon metabolism. International Journal of Molecular Sciences, 19(10). https://doi.org/10.3390/ijms19103106.
Mahmoud, A. M., & Ali, M. M. (2019). Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients, 11(3). https://doi.org/10.3390/nu11030608.
Gut, P., & Verdin, E. (2013). The nexus of chromatin regulation and intermediary metabolism. Nature, 502(7472), 489–498. https://doi.org/10.1038/nature12752.
Kaelin Jr., W. G., & McKnight, S. L. (2013). Influence of metabolism on epigenetics and disease. Cell, 153(1), 56–69. https://doi.org/10.1016/j.cell.2013.03.004.
Xiao, M., Yang, H., Xu, W., Ma, S., Lin, H., Zhu, H., et al. (2012). Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & Development, 26(12), 1326–1338. https://doi.org/10.1101/gad.191056.112.
Xu, Y., Wu, F., Tan, L., Kong, L., Xiong, L., Deng, J., et al. (2011). Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular Cell, 42(4), 451–464. https://doi.org/10.1016/j.molcel.2011.04.005.
Flavahan, W. A., Drier, Y., Liau, B. B., Gillespie, S. M., Venteicher, A. S., Stemmer-Rachamimov, A. O., et al. (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature, 529(7584), 110–114. https://doi.org/10.1038/nature16490.
Cimmino, L., Neel, B. G., & Aifantis, I. (2018). Vitamin C in stem cell reprogramming and cancer. Trends in Cell Biology, 28(9), 698–708. https://doi.org/10.1016/j.tcb.2018.04.001.
Aoki, K., & Natsume, A. (2019). Overview of DNA methylation in adult diffuse gliomas. Brain Tumor Pathology, 36(2), 84–91. https://doi.org/10.1007/s10014-019-00339-w.
Bouras, E., Karakioulaki, M., Bougioukas, K. I., Aivaliotis, M., Tzimagiorgis, G., & Chourdakis, M. (2019). Gene promoter methylation and cancer: an umbrella review. Gene, 710, 333–340. https://doi.org/10.1016/j.gene.2019.06.023.
Roll, J. D., Rivenbark, A. G., Sandhu, R., Parker, J. S., Jones, W. D., Carey, L. A., et al. (2013). Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Experimental and Molecular Pathology, 95(3), 276–287. https://doi.org/10.1016/j.yexmp.2013.09.001.
Phuong, N. T., Kim, S. K., Lim, S. C., Kim, H. S., Kim, T. H., Lee, K. Y., et al. (2011). Role of PTEN promoter methylation in tamoxifen-resistant breast cancer cells. Breast Cancer Research and Treatment, 130(1), 73–83. https://doi.org/10.1007/s10549-010-1304-2.
Yang, L., Yu, S. J., Hong, Q., Yang, Y., & Shao, Z. M. (2015). Reduced expression of TET1, TET2, TET3 and TDG mRNAs are associated with poor prognosis of patients with early breast cancer. PLoS One, 10(7), e0133896. https://doi.org/10.1371/journal.pone.0133896.
Thienpont, B., Galle, E., & Lambrechts, D. (2016). TET enzymes as oxygen-dependent tumor suppressors: exciting new avenues for cancer management. Epigenomics, 8(11), 1445–1448. https://doi.org/10.2217/epi-2016-0126.
Kao, S. H., Wu, K. J., & Lee, W. H. (2016). Hypoxia, epithelial-mesenchymal transition, and TET-mediated epigenetic changes. Journal of Clinical Medicine, 5(2). https://doi.org/10.3390/jcm5020024.
Adam, J., Yang, M., Soga, T., & Pollard, P. J. (2014). Rare insights into cancer biology. Oncogene, 33(20), 2547–2556. https://doi.org/10.1038/onc.2013.222.
Laukka, T., Mariani, C. J., Ihantola, T., Cao, J. Z., Hokkanen, J., Kaelin Jr., W. G., et al. (2016). Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. The Journal of Biological Chemistry, 291(8), 4256–4265. https://doi.org/10.1074/jbc.M115.688762.
Liu, Y., Jiang, W., Liu, J., Zhao, S., Xiong, J., Mao, Y., et al. (2012). IDH1 mutations inhibit multiple alpha-ketoglutarate-dependent dioxygenase activities in astroglioma. Journal of Neuro-Oncology, 109(2), 253–260. https://doi.org/10.1007/s11060-012-0914-4.
Agnihotri, S., Aldape, K. D., & Zadeh, G. (2014). Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma. Neurosurgical Focus, 37(6), E13. https://doi.org/10.3171/2014.9.Focus14505.
Budczies, J., & Denkert, C. (2016). Tissue-based metabolomics to analyze the breast cancer metabolome. Recent Results in Cancer Research, 207, 157–175. https://doi.org/10.1007/978-3-319-42118-6_7.
Hervouet, E., Peixoto, P., Delage-Mourroux, R., Boyer-Guittaut, M., & Cartron, P. F. (2018). Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clinical Epigenetics, 10, 17. https://doi.org/10.1186/s13148-018-0450-y.
Inoue, S., Li, W. Y., Tseng, A., Beerman, I., Elia, A. J., Bendall, S. C., et al. (2016). Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell, 30(2), 337–348. https://doi.org/10.1016/j.ccell.2016.05.018.
Yan, H., Parsons, D. W., Jin, G., McLendon, R., Rasheed, B. A., Yuan, W., et al. (2009). IDH1 and IDH2 mutations in gliomas. The New England Journal of Medicine, 360(8), 765–773. https://doi.org/10.1056/NEJMoa0808710.
Muller, T., Gessi, M., Waha, A., Isselstein, L. J., Luxen, D., Freihoff, D., et al. (2012). Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. The American Journal of Pathology, 181(2), 675–683. https://doi.org/10.1016/j.ajpath.2012.04.017.
Turcan, S., Fabius, A. W., Borodovsky, A., Pedraza, A., Brennan, C., Huse, J., et al. (2013). Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT inhibitor decitabine. Oncotarget, 4(10), 1729–1736. https://doi.org/10.18632/oncotarget.1412.
Zhou, Z., Li, H. Q., & Liu, F. (2018). DNA methyltransferase inhibitors and their therapeutic potential. Current Topics in Medicinal Chemistry, 18(28), 2448–2457. https://doi.org/10.2174/1568026619666181120150122.
Chen, A., Sceneay, J., Godde, N., Kinwel, T., Ham, S., Thompson, E. W., et al. (2018). Intermittent hypoxia induces a metastatic phenotype in breast cancer. Oncogene, 37(31), 4214–4225. https://doi.org/10.1038/s41388-018-0259-3.
Flavahan, W. A., Gaskell, E., & Bernstein, B. E. (2017). Epigenetic plasticity and the hallmarks of cancer. Science, 357(6348). https://doi.org/10.1126/science.aal2380.
Prasad, P., Mittal, S. A., Chongtham, J., Mohanty, S., & Srivastava, T. (2017). Hypoxia-mediated epigenetic regulation of stemness in brain tumor cells. Stem Cells, 35(6), 1468–1478. https://doi.org/10.1002/stem.2621.
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature, 367(6464), 645–648. https://doi.org/10.1038/367645a0.
Reya, T., Morrison, S. J., Clarke, M. F., & Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature, 414(6859), 105–111. https://doi.org/10.1038/35102167.
Giordano, A., Gao, H., Cohen, E. N., Anfossi, S., Khoury, J., Hess, K., et al. (2013). Clinical relevance of cancer stem cells in bone marrow of early breast cancer patients. Annals of oncology : official journal of the European Society for Medical Oncology, 24(10), 2515–2521. https://doi.org/10.1093/annonc/mdt223.
Bu, Y., & Cao, D. (2012). The origin of cancer stem cells. Frontiers in Bioscience (Scholar Edition), 4, 819–830. https://doi.org/10.2741/s302.
Prieto-Vila, M., Takahashi, R.-U., Usuba, W., Kohama, I., & Ochiya, T. (2017). Drug resistance driven by cancer stem cells and their niche. International Journal of Molecular Sciences, 18(12). https://doi.org/10.3390/ijms18122574.
Lobo, N. A., Shimono, Y., Qian, D., & Clarke, M. F. (2007). The biology of cancer stem cells. Annual Review of Cell and Developmental Biology, 23(1), 675–699. https://doi.org/10.1146/annurev.cellbio.22.010305.104154.
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., & Clarke, M.F. F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA, 100(7), 3983–3988
Hwang-Verslues, W. W., Kuo, W.-H., Chang, P.-H., Pan, C.-C., Wang, H.-H., Tsai, S.-T., et al. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem. Cell Markers. PLoS ONE, 4(12), 8377–8377. https://doi.org/10.1371/journal.pone.0008377.
Figueiredo, E., Zaghloul, K., Zhang, Y., Tan, S. T., Bradshaw, A., Wickremsekera, A., et al. (2016). Cancer stem cell hierarchy in glioblastoma multiforme. 3, 2, doi:https://doi.org/10.3389/fsurg.2016.00021.
Bliss, S. A., Sinha, G., Sandiford, O. A., Williams, L. M., Engelberth, D. J., Guiro, K., et al. (2016). Mesenchymal stem cell–derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Research, 76(19), 5832–5844. https://doi.org/10.1158/0008-5472.CAN-16-1092.
Mani, S. A., Guo, W., Liao, M.-J., Eaton, E. N., Ayyanan, A., Zhou, A. Y., et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell, 133(4), 704–715. https://doi.org/10.1016/j.cell.2008.03.027.
Sell, S. (2004). Stem cell origin of cancer and differentiation therapy. Critical Reviews in Oncology/Hematology, 51(1), 1–28. https://doi.org/10.1016/J.CRITREVONC.2004.04.007.
Wicha, M. S., Liu, S., & Dontu, G. (2006). Cancer stem cells: an old idea-A paradigm shift. Cancer Research, 66(4), 1883–1890. https://doi.org/10.1158/0008-5472.CAN-05-3153.
Marsden, C. G., Wright, M. J., Carrier, L., Moroz, K., & Rowan, B. G. (2012). Disseminated breast cancer cells acquire a highly malignant and aggressive metastatic phenotype during metastatic latency in the bone. PLoS One, 7(11), e47587–e47587. https://doi.org/10.1371/journal.pone.0047587.
Shiozawa, Y., & Taichman, R. S. (2012). Cancer stem cells and the bone marrow microenvironment. Bonekey Reports, 2012(1). https://doi.org/10.1038/bonekey.2012.48.
Birbrair, A., & Frenette, P. S. (2016). Niche heterogeneity in the bone marrow. Annals of the New York Academy of Sciences, 1370(1), 82–96. https://doi.org/10.1111/nyas.13016.
Morrison, S. J., & Scadden, D. T. (2014). The bone marrow niche for haematopoietic stem cells. Nature, 505(7483), 327–334. https://doi.org/10.1038/nature12984.
Yu, V. W., & Scadden, D. T. (2016). Hematopoietic stem cell and its bone marrow niche. Current Topics in Developmental Biology, 118, 21–44. https://doi.org/10.1016/bs.ctdb.2016.01.009.
Guerra, D. A. P., Paiva, A. E., Sena, I. F. G., Azevedo, P. O., Batista Jr., M. L., Mintz, A., et al. (2018). Adipocytes role in the bone marrow niche. Cytometry. Part A, 93(2), 167–171. https://doi.org/10.1002/cyto.a.23301.
Rameshwar, P. (2010). Breast cancer cell dormancy in bone marrow: potential therapeutic targets within the marrow microenvironment. Expert Review of Anticancer Therapy, 10(2), 129–132. https://doi.org/10.1586/era.10.3.
Eltoukhy, H. S., Sinha, G., Moore, C. A., Gergues, M., & Rameshwar, P. (2018). Secretome within the bone marrow microenvironment: a basis for mesenchymal stem cell treatment and role in cancer dormancy. Biochimie, 155, 92–103. https://doi.org/10.1016/j.biochi.2018.05.018.
Ridge, S. M., Sullivan, F. J., & Glynn, S. A. (2017). Mesenchymal stem cells: key players in cancer progression. Molecular Cancer, 16(1), 31. https://doi.org/10.1186/s12943-017-0597-8.
Ruivo, C. F., Adem, B., Silva, M., & Melo, S. A. (2017). The biology of cancer exosomes: insights and new perspectives. Cancer Research, 77(23), 6480–6488. https://doi.org/10.1158/0008-5472.CAN-17-0994.
Walker, N. D., Elias, M., Guiro, K., Bhatia, R., Greco, S. J., Bryan, M., et al. (2019). Exosomes from differentially activated macrophages influence dormancy or resurgence of breast cancer cells within bone marrow stroma. Cell Death & Disease, 10(2), 59. https://doi.org/10.1038/s41419-019-1304-z.
Wu, J. I., & Wang, L. H. (2019). Emerging roles of gap junction proteins connexins in cancer metastasis, chemoresistance and clinical application. Journal of Biomedical Science, 26(1), 8. https://doi.org/10.1186/s12929-019-0497-x.
Zefferino, R., Piccoli, C., Gioia, S. D., Capitanio, N., & Conese, M. (2019). Gap junction intercellular communication in the carcinogenesis hallmarks: is this a phenomenon or epiphenomenon? Cells, 8(8). https://doi.org/10.3390/cells8080896.
Sinyuk, M., Mulkearns-Hubert, E. E., Reizes, O., & Lathia, J. (2018). Cancer connectors: connexins, gap junctions, and communication. Frontiers in Oncology, 8, 646. https://doi.org/10.3389/fonc.2018.00646.
Busby, M., Hallett, M. T., & Plante, I. (2018). The complex subtype-dependent role of connexin 43 (GJA1) in breast cancer. International Journal of Molecular Sciences, 19(3). https://doi.org/10.3390/ijms19030693.
Oliveira, R., Christov, C., Guillamo, J. S., de Bouard, S., Palfi, S., Venance, L., et al. (2005). Contribution of gap junctional communication between tumor cells and astroglia to the invasion of the brain parenchyma by human glioblastomas. BMC Cell Biology, 6(1), 7. https://doi.org/10.1186/1471-2121-6-7.
Munoz, J. L., Bliss, S. A., Greco, S. J., Ramkissoon, S. H., Ligon, K. L., & Rameshwar, P. (2013). Delivery of functional anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Molecular Therapy--Nucleic Acids, 2, e126. https://doi.org/10.1038/mtna.2013.60.
Park, J. M., Munoz, J. L., Won, B. W., Bliss, S. A., Greco, S. J., Patel, S. A., et al. (2013). Exogenous CXCL12 activates protein kinase C to phosphorylate connexin 43 for gap junctional intercellular communication among confluent breast cancer cells. Cancer Letters, 331(1), 84–91. https://doi.org/10.1016/j.canlet.2012.12.007.
Poddar, S., Kesharwani, D., & Datta, M. (2017). Interplay between the miRNome and the epigenetic machinery: implications in health and disease. Journal of Cellular Physiology, 232(11), 2938–2945. https://doi.org/10.1002/jcp.25819.
Piletic, K., & Kunej, T. (2016). MicroRNA epigenetic signatures in human disease. Archives of Toxicology, 90(10), 2405–2419. https://doi.org/10.1007/s00204-016-1815-7.
Tu, J., Ng, S. H., Luk, A. C., Liao, J., Jiang, X., Feng, B., et al. (2015). MicroRNA-29b/Tet1 regulatory axis epigenetically modulates mesendoderm differentiation in mouse embryonic stem cells. Nucleic Acids Research, 43(16), 7805–7822. https://doi.org/10.1093/nar/gkv653.
Iwama, H., Kato, K., Imachi, H., Murao, K., & Masaki, T. (2018). Human microRNAs preferentially target genes with intermediate levels of expression and its formation by mammalian evolution. PLoS One, 13(5), e0198142. https://doi.org/10.1371/journal.pone.0198142.
Liu, X., Chen, X., Yu, X., Tao, Y., Bode, A. M., Dong, Z., et al. (2013). Regulation of microRNAs by epigenetics and their interplay involved in cancer. Journal of Experimental & Clinical Cancer Research, 32, 96. https://doi.org/10.1186/1756-9966-32-96.
Daniunaite, K., Dubikaityte, M., Gibas, P., Bakavicius, A., Rimantas Lazutka, J., Ulys, A., et al. (2017). Clinical significance of miRNA host gene promoter methylation in prostate cancer. Human Molecular Genetics, 26(13), 2451–2461. https://doi.org/10.1093/hmg/ddx138.
Pathania, R. a. R. S. a. E. S. a. P. R. a. Y. P. a. C. S. a. V.-K. R. a. A. P. a. G.-P. J. P. a. S. (2015). DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nature Communications, 6(1), 6910. https://doi.org/10.1038/ncomms7910.
Oltra, S. S., Peña-Chilet, M., Flower, K., Martinez, M. T., Alonso, E., Burgues, O., et al. (2019). Acceleration in the DNA methylation age in breast cancer tumours from very young women. Scic Rep, 9(1). https://doi.org/10.1038/s41598-019-51457-6.
El Helou, R., Wicinski, J., Guille, A., Adélaïde, J., Finetti, P., Bertucci, F., et al. (2014). Brief reports: a distinct DNA methylation signature defines breast cancer stem cells and predicts cancer outcome. Stem Cells, 32(11), 3031–3036. https://doi.org/10.1002/stem.1792.
Taube, J. H., Malouf, G. G., Lu, E., Sphyris, N., Vijay, V., Ramachandran, P. P., et al. (2013). Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Scientific Reports, 3, 2687. https://doi.org/10.1038/srep02687.
Neves, R., Scheel, C., Weinhold, S., Honisch, E., Iwaniuk, K. M., Trompeter, H. I., et al. (2010). Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Research Notes, 3, 219. https://doi.org/10.1186/1756-0500-3-219.
Nass, S. J., Herman, J. G., Gabrielson, E., Iversen, P. W., Parl, F. F., Davidson, N. E., et al. (2000). Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Research, 60(16), 4346–4348.
Joo, J. E., Dowty, J. G., Milne, R. L., Wong, E. M., Dugue, P. A., English, D., et al. (2018). Heritable DNA methylation marks associated with susceptibility to breast cancer. Nature Communications, 9(1), 867. https://doi.org/10.1038/s41467-018-03058-6.
Fleischer, T., Tekpli, X., Mathelier, A., Wang, S., Nebdal, D., Dhakal, H. P., et al. (2017). DNA methylation at enhancers identifies distinct breast cancer lineages. Nature Communications, 8, 1379. https://doi.org/10.1038/s41467-017-00510-x.
Hernandez-Vargas, H., Ouzounova, M., Le Calvez-Kelm, F., Lambert, M. P., McKay-Chopin, S., Tavtigian, S. V., et al. (2011). Methylome analysis reveals Jak-STAT pathway deregulation in putative breast cancer stem cells. Epigenetics, 6(4), 428–439. https://doi.org/10.4161/epi.6.4.14515.
Chimonidou, M., Strati, A., Tzitzira, A., Sotiropoulou, G., Malamos, N., Georgoulias, V., et al. (2011). DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clinical Chemistry, 57(8), 1169–1177. https://doi.org/10.1373/clinchem.2011.165902.
Darvin, P., Sasidharan Nair, V., & Elkord, E. (2019). PD-L1 expression in human breast cancer stem cells is epigenetically regulated through posttranslational histone modifications. Journal of Oncology, 2019, 3958908. https://doi.org/10.1155/2019/3958908.
Rankin, E. B., & Giaccia, A. J. (2008). The role of hypoxia-inducible factors in tumorigenesis. Cell Death and Differentiation, 15(4), 678–685. https://doi.org/10.1038/cdd.2008.21.
Guo, C., Chen, L. H., Huang, Y., Chang, C. C., Wang, P., Pirozzi, C. J., et al. (2013). KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget, 4(11), 2144–2153. https://doi.org/10.18632/oncotarget.1555.
Wang, L., Ozark, P. A., Smith, E. R., Zhao, Z., Marshall, S. A., Rendleman, E. J., et al. (2018). TET2 coactivates gene expression through demethylation of enhancers. Science Advances, 4(11), eaau6986. https://doi.org/10.1126/sciadv.aau6986.
Chen, J. Y., Luo, C. W., Lai, Y. S., Wu, C. C., & Hung, W. C. (2017). Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis, 6(8), e369. https://doi.org/10.1038/oncsis.2017.71.
Yu, Y., Qi, J., Xiong, J., Jiang, L., Cui, D., He, J., et al. (2019). Epigenetic co-deregulation of EZH2/TET1 is a senescence-countering, actionable vulnerability in triple-negative breast cancer. Theranostics, 9(3), 761–777. https://doi.org/10.7150/thno.29520.
Wu, M. Z., Chen, S. F., Nieh, S., Benner, C., Ger, L. P., Jan, C. I., et al. (2015). Hypoxia drives breast tumor malignancy through a TET-TNFalpha-p38-MAPK signaling axis. Cancer Research, 75(18), 3912–3924. https://doi.org/10.1158/0008-5472.CAN-14-3208.
Thienpont, B., Steinbacher, J., Zhao, H., D’Anna, F., Kuchnio, A., Ploumakis, A., et al. (2016). Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature, 537(7618), 63–68. https://doi.org/10.1038/nature19081.
Ryan Good, C., Panjarian, S., Kelly, A. D., Madzo, J., Patel, B., Jelinek, J., et al. (2018). Genome and epigenome TET1-mediated hypomethylation activates oncogenic signaling in triple-negative. Breast Cancer. https://doi.org/10.1158/0008-5472.CAN-17-2082.
Hsu, C. H., Peng, K. L., Kang, M. L., Chen, Y. R., Yang, Y. C., Tsai, C. H., et al. (2012). TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Reports, 2(3), 568–579. https://doi.org/10.1016/j.celrep.2012.08.030.
Collignon, E., Canale, A., Al Wardi, C., Bizet, M., Calonne, E., Dedeurwaerder, S., et al. (2018). Immunity drives TET1 regulation in cancer through NF-kappaB. Science Advances, 4(6), eaap7309. https://doi.org/10.1126/sciadv.aap7309.
Sant, D. W., Mustafi, S., Gustafson, C. B., Chen, J., Slingerland, J. M., & Wang, G. (2018). Vitamin C promotes apoptosis in breast cancer cells by increasing TRAIL expression. Scientific Reports, 8(1), 5306. https://doi.org/10.1038/s41598-018-23714-7.
Klughammer, J., Kiesel, B., Roetzer, T., Fortelny, N., Nemc, A., Nenning, K. H., et al. (2018). The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nature Medicine, 24(10), 1611–1624. https://doi.org/10.1038/s41591-018-0156-x.
Lee, E. J., Rath, P., Liu, J., Ryu, D., Pei, L., Noonepalle, S. K., et al. (2015). Identification of global DNA methylation signatures in glioblastoma-derived cancer stem cells. Journal of Genetics and Genomics, 42(7), 355–371. https://doi.org/10.1016/j.jgg.2015.06.003.
Bulstrode, H., Johnstone, E., Marques-Torrejon, M. A., Ferguson, K. M., Bressan, R. B., Blin, C., et al. (2017). Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes & Development, 31(8), 757–773. https://doi.org/10.1101/gad.293027.116.
Gopisetty, G., Xu, J., Sampath, D., Colman, H., & Puduvalli, V. K. (2013). Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene, 32(26), 3119–3129. https://doi.org/10.1038/onc.2012.331.
Iwadate, Y., Suganami, A., Tamura, Y., Matsutani, T., Hirono, S., Shinozaki, N., et al. (2017). The pluripotent stem-cell marker alkaline phosphatase is highly expressed in refractory glioblastoma with DNA hypomethylation. Neurosurgery, 80(2), 248–256. https://doi.org/10.1093/neuros/nyw026.
Zhou, D., Wan, Y., Xie, D., Wang, Y., Wei, J., Yan, Q., et al. (2015). DNMT1 mediates chemosensitivity by reducing methylation of miRNA-20a promoter in glioma cells. Experimental & Molecular Medicine, 47, e182. https://doi.org/10.1038/emm.2015.57.
Ning, X., Shi, Z., Liu, X., Zhang, A., Han, L., Jiang, K., et al. (2015). DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Letters, 359(2), 198–205. https://doi.org/10.1016/j.canlet.2015.01.005.
Hervouet, E., Vallette, F. M., & Cartron, P. F. (2010). Impact of the DNA methyltransferases expression on the methylation status of apoptosis-associated genes in glioblastoma multiforme. Cell Death & Disease, 1, e8. https://doi.org/10.1038/cddis.2009.7.
Gu, X., Gong, H., Shen, L., & Gu, Q. (2018). MicroRNA-129-5p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting DNMT3A. American Journal of Translational Research, 10(9), 2834–2847.
Sun, J., Tian, X., Zhang, J., Huang, Y., Lin, X., Chen, L., et al. (2017). Regulation of human glioma cell apoptosis and invasion by miR-152-3p through targeting DNMT1 and regulating NF2 : MiR-152-3p regulate glioma cell apoptosis and invasion. Journal of Experimental & Clinical Cancer Research, 36(1), 100. https://doi.org/10.1186/s13046-017-0567-4.
Izumikawa, K., Ishikawa, H., Simpson, R. J., & Takahashi, N. (2018). Modulating the expression of Chtop, a versatile regulator of gene-specific transcription and mRNA export. RNA Biology, 15(7), 849–855. https://doi.org/10.1080/15476286.2018.1465795.
Johnson, K. C., Houseman, E. A., King, J. E., von Herrmann, K. M., Fadul, C. E., & Christensen, B. C. (2016). 5-Hydroxymethylcytosine localizes to enhancer elements and is associated with survival in glioblastoma patients. Nature Communications, 7, 13177. https://doi.org/10.1038/ncomms13177.
Carella, A., Tejedor, J. R., García, M. G., Urdinguio, R. G., Bayón, G. F., Sierra, M., et al. (2020). Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. International Journal of Cancer, 146(2), 373–387. https://doi.org/10.1002/ijc.32520.
Qu, Q., Sun, G., Li, W., Yang, S., Ye, P., Zhao, C., et al. (2010). Orphan nuclear receptor TLX activates Wnt/Β-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nature Cell Biology, 12(1), 31–40. https://doi.org/10.1038/ncb2001.
Cui, Q., Yang, S., Ye, P., Tian, E., Sun, G., Zhou, J., et al. (2016). Downregulation of TLX induces TET3 expression and inhibits glioblastoma stem cell self-renewal and tumorigenesis. Nature Communications, 7, 10637. https://doi.org/10.1038/ncomms10637.
Zhou, D., Alver, B. M., Li, S., Hlady, R. A., Thompson, J. J., Schroeder, M. A., et al. (2018). Distinctive epigenomes characterize glioma stem cells and their response to differentiation cues. Genome Biology, 19(1), 43–43. https://doi.org/10.1186/s13059-018-1420-6.
Puig, I., Tenbaum, S. P., Chicote, I., Arques, O., Martinez-Quintanilla, J., Cuesta-Borras, E., et al. (2018). TET2 controls chemoresistant slow-cycling cancer cell survival and tumor recurrence. The Journal of Clinical Investigation, 128(9), 3887–3905. https://doi.org/10.1172/jci96393.
Garcia, M. G., Carella, A., Urdinguio, R. G., Bayon, G. F., Lopez, V., Tejedor, J. R., et al. (2018). Epigenetic dysregulation of TET2 in human glioblastoma. Oncotarget, 9(40), 25922–25934. https://doi.org/10.18632/oncotarget.25406.
Huang, Y., Chavez, L., Chang, X., Wang, X., Pastor, W. A., Kang, J., et al. (2014). Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America, 111(4), 1361–1366. https://doi.org/10.1073/pnas.1322921111.
Xu, Y., Chao, L., Wang, J., & Sun, Y. (2017). miRNA-148a regulates the expression of the estrogen receptor through DNMT1-mediated DNA methylation in breast cancer cells. Oncology Letters, 14(4), 4736–4740. https://doi.org/10.3892/ol.2017.6803.
Cho, J. H., Dimri, M., & Dimri, G. P. (2015). MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. The Journal of Biological Chemistry, 290(16), 10555–10567. https://doi.org/10.1074/jbc.M114.624361.
Song, J., Jin, E. H., Kim, D., Kim, K. Y., Chun, C. H., & Jin, E. J. (2015). MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during osteoarthritis pathogenesis. BBA Clinical, 3, 79–89. https://doi.org/10.1016/j.bbacli.2014.11.009.
Jiang, S. (2019). MicroRNA Let-7adf in Tet regulation. Aging, 11(14), 4772–4773. https://doi.org/10.18632/aging.102101.
Fu, X., Jin, L., Wang, X., Luo, A., Hu, J., Zheng, X., et al. (2013). MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proceedings of the National Academy of Sciences of the United States of America, 110(44), 17892–17897. https://doi.org/10.1073/pnas.1317397110.
Cheng, J., Guo, S., Chen, S., Mastriano, S. J., Liu, C., D’Alessio, A. C., et al. (2013). An extensive network of TET2-targeting microRNAs regulates malignant hematopoiesis. Cell Reports, 5(2), 471–481. https://doi.org/10.1016/j.celrep.2013.08.050.
Yang, H., Li, Q., Zhao, W., Yuan, D., Zhao, H., & Zhou, Y. (2014). miR-329 suppresses the growth and motility of neuroblastoma by targeting KDM1A. FEBS Letters, 588(1), 192–197. https://doi.org/10.1016/j.febslet.2013.11.036.
Nilsson, E. M., Laursen, K. B., Whitchurch, J., McWilliam, A., Odum, N., Persson, J. L., et al. (2015). MiR137 is an androgen regulated repressor of an extended network of transcriptional coregulators. Oncotarget, 6(34), 35710–35725. https://doi.org/10.18632/oncotarget.5958.
Bao, J., Zou, J. H., Li, C. Y., & Zheng, G. Q. (2016). miR-194 inhibits gastric cancer cell proliferation and tumorigenesis by targeting KDM5B. European Review for Medical and Pharmacological Sciences, 20(21), 4487–4493.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ferrer, A.I., Trinidad, J.R., Sandiford, O. et al. Epigenetic dynamics in cancer stem cell dormancy. Cancer Metastasis Rev 39, 721–738 (2020). https://doi.org/10.1007/s10555-020-09882-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-020-09882-x