Abstract
Purpose
Immunohistochemistry (IHC) assessment of the estrogen receptor (ER) status has low consensus among pathologists. Quantitative transcriptional signatures are highly sensitive to the measurement variation and sample quality. Here, we developed a robust qualitative signature, based on within-sample relative expression orderings (REOs) of genes, to reclassify ER status.
Methods
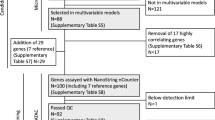
From the gene pairs with significantly stable REOs in ER+ samples and reversely stable REOs in ER− samples, concordantly identified from four datasets, we extracted a signature to determine a sample’s ER status through evaluating whether the REOs within the sample significantly match with the ER+ REOs or the ER− REOs.
Results
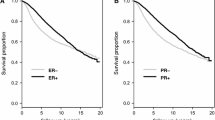
A signature with 112 gene pairs was extracted. It was validated through evaluating whether the reclassified ER+ or ER− patients could benefit from tamoxifen therapy or neoadjuvant chemotherapy. In three datasets for IHC-determined ER+ patients treated with post-operative tamoxifen therapy, 11.6–12.4% patients were reclassified as ER− by the signature and, as expected, they had significantly worse recurrence-free survival than the ER+ patients confirmed by the signature. On another hand, in two datasets for IHC-determined ER− patients treated with neoadjuvant chemotherapy, 18.8 and 7.8% patients were reclassified as ER+ and, as expected, their pathological complete response rate was significantly lower than that of the other ER− patients confirmed by the signature.
Conclusions
The REO-based signature can provide an objective assessment of ER status of breast cancer patients and effectively reduce misjudgments of ER status by IHC.


Similar content being viewed by others
Abbreviations
- ER:
-
Estrogen receptor
- IHC:
-
Immunohistochemical
- REO:
-
Relative expression ordering
- pCR:
-
Pathological complete response
- RD:
-
Residual disease
- GEO:
-
Gene expression omnibus
- RMA:
-
Robust multichip average algorithm
- RFS:
-
Relapse-free survival
- HR:
-
Hazard ratio
- CI:
-
Confidence interval
- FDR:
-
False discovery rate
References
Jemal A, Bray F, Center MM et al (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90
American Cancer Society. Available from: https://www.cancer.org/cancer/breast-cancer.html
Gancberg D, Jarvinen T, di Leo A et al (2002) Evaluation of HER-2/NEU protein expression in breast cancer by immunohistochemistry: an interlaboratory study assessing the reproducibility of HER-2/NEU testing. Breast Cancer Res Treat 74(2):113–120
Diaz LK, Sahin A, Sneige N (2004) Interobserver agreement for estrogen receptor immunohistochemical analysis in breast cancer: a comparison of manual and computer-assisted scoring methods. Ann Diagn Pathol 8(1):23–27
Kirkegaard T, Edwards J, Tovey S et al (2006) Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology 48(7):787–794
Arihiro K, Umemura S, Kurosumi M et al (2007) Comparison of evaluations for hormone receptors in breast carcinoma using two manual and three automated immunohistochemical assays. Am J Clin Pathol 127(3):356–365
Press MF, Slamon DJ, Flom KJ et al (2002) Evaluation of HER-2/neu gene amplification and overexpression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J Clin Oncol 20(14):3095–3105
Fitzgibbons PL, Murphy DA, Hammond ME et al (2010) Recommendations for validating estrogen and progesterone receptor immunohistochemistry assays. Arch Pathol Lab Med 134(6):930–935
Hammond ME, Hayes DF, Dowsett M et al (2010) American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28(16):2784–2795
Orlando L, Viale G, Bria E et al (2016) Discordance in pathology report after central pathology review: implications for breast cancer adjuvant treatment. Breast 30:151–155
Dubowitz V (1991) A new muscle journal for the nineties. Neuromuscul Disord: NMD 1(1):1–2
Sheffield BS, Kos Z, Asleh-Aburaya K et al (2016) Molecular subtype profiling of invasive breast cancers weakly positive for estrogen receptor. Breast Cancer Res Treat 155(3):483–490
Gong Y, Yan K, Lin F et al (2007) Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: a gene-expression profiling study. Lancet Oncol 8(3):203–211
Badve SS, Baehner FL, Gray RP et al (2008) Estrogen- and progesterone-receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol 26(15):2473–2481
Roepman P, Horlings HM, Krijgsman O et al (2009) Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin Cancer Res 15(22):7003–7011
Du X, Li XQ, Li L et al (2013) The detection of ESR1/PGR/ERBB2 mRNA levels by RT-QPCR: a better approach for subtyping breast cancer and predicting prognosis. Breast Cancer Res Treat 138(1):59–67
Tramm T, Hennig G, Kyndi M et al (2013) Reliable PCR quantitation of estrogen, progesterone and ERBB2 receptor mRNA from formalin-fixed, paraffin-embedded tissue is independent of prior macro-dissection. Virchows Archiv 463(6):775–786
Wilson TR, Xiao Y, Spoerke JM et al (2014) Development of a robust RNA-based classifier to accurately determine ER, PR, and HER2 status in breast cancer clinical samples. Breast Cancer Res Treat 148(2):315–325
Consortium M, Shi L, Reid LH et al (2006) The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 24(9):1151–1161
Gazdar AF, Schiller JH (2011) Predictive and prognostic factors for non-small cell lung cancer–potholes in the road to the promised land. J Natl Cancer Inst 103(24):1810–1811
Qi L, Chen L, Li Y et al (2016) Critical limitations of prognostic signatures based on risk scores summarized from gene expression levels: a case study for resected stage I non-small-cell lung cancer. Brief Bioinform 17(2):233–242
Lu X, Lu X, Wang ZC et al (2008) Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat 108(2):191–201
Wang D, Cheng L, Wang M et al (2011) Extensive increase of microarray signals in cancers calls for novel normalization assumptions. Comput Biol Chem 35(3):126–130
Nygaard V, Rodland EA, Hovig E (2016) Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 17(1):29–39
Eddy JA, Sung J, Geman D et al (2010) Relative expression analysis for molecular cancer diagnosis and prognosis. Technol Cancer Res Treat 9(2):149–159
Patil P, Bachant-Winner PO, Haibe-Kains B et al (2015) Test set bias affects reproducibility of gene signatures. Bioinformatics 31(14):2318–2323
Cheng J, Guo Y, Gao Q et al (2017) Circumvent the uncertainty in the applications of transcriptional signatures to tumor tissues sampled from different tumor sites. Oncotarget 8(18):30265–30275
Chen R, Guan Q, Cheng J et al (2017) Robust transcriptional tumor signatures applicable to both formalin-fixed paraffin-embedded and fresh-frozen samples. Oncotarget 8(4):6652–6662
Liu H, Li Y, He J et al (2017) Robust transcriptional signatures for low-input RNA samples based on relative expression orderings. BMC Genom 18(1):913
Ao L, Song X, Li X et al (2016) An individualized prognostic signature and multiomics distinction for early stage hepatocellular carcinoma patients with surgical resection. Oncotarget 7(17):24097–24110
Li X, Cai H, Zheng W et al (2016) An individualized prognostic signature for gastric cancer patients treated with 5-Fluorouracil-based chemotherapy and distinct multi-omics characteristics of prognostic groups. Oncotarget 7(8):8743–8755
Qi L, Li T, Shi G et al (2017) An individualized gene expression signature for prediction of lung adenocarcinoma metastases. Mol Oncol 11(11):1630–1645
Guan Q, Chen R, Yan H et al (2016) Differential expression analysis for individual cancer samples based on robust within-sample relative gene expression orderings across multiple profiling platforms. Oncotarget 7(42):68909–68920
Mouttet D, Lae M, Caly M et al (2016) Estrogen-receptor, progesterone-receptor and HER2 status determination in invasive breast cancer. Concordance between immuno-histochemistry and MapQuant microarray based assay. PLoS ONE 11(2):e0146474
Wesseling J, Tinterri C, Sapino A et al (2016) An international study comparing conventional versus mRNA level testing (TargetPrint) for ER, PR, and HER2 status of breast cancer. Virchows Archiv 469(3):297–304
Viale G, de Snoo FA, Slaets L et al (2017) Immunohistochemical versus molecular (BluePrint and MammaPrint) subtyping of breast carcinoma. Outcome results from the EORTC 10041/BIG 3-04 MINDACT trial. Breast Cancer Res Treat
Early Breast Cancer Trialists’ Collaborative G, Davies C, Godwin J et al (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378(9793):771–784
Hackshaw A, Roughton M, Forsyth S et al (2011) Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol 29(13):1657–1663
Blamey RW, Bates T, Chetty U et al (2013) Radiotherapy or tamoxifen after conserving surgery for breast cancers of excellent prognosis: British Association of Surgical Oncology (BASO) II trial. Eur J Cancer 49(10):2294–2302
Davies C, Pan H, Godwin J et al (2013) Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 381(9869):805–816
Guarneri V, Broglio K, Kau SW et al (2006) Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol 24(7):1037–1044
Kaufmann M, von Minckwitz G, Smith R et al (2003) International expert panel on the use of primary (preoperative) systemic treatment of operable breast cancer: review and recommendations. J Clin Oncol 21(13):2600–2608
Irizarry RA, Hobbs B, Collin F et al (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4(2):249–264
Bahn AK (1969) Application of binomial distribution to medicine: comparison of one sample proportion to an expected proportion (for small samples). Evaluation of a new treatment. Evaluation of a risk factor. J Am Med Women’s Assoc 24(12):957–966
Schweder T, Spjøtvoll E (1982) A class of rank test procedures for censored survival data. Biometrika 69(3):553–566
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc 57(1):289–300
National Comprehensive Cancer Network. Available from: https://www.nccn.org/professionals/physician_gls/default.aspx
Yi M, Huo L, Koenig KB et al (2014) Which threshold for ER positivity? A retrospective study based on 9639 patients. Ann Oncol 25(5):1004–1011
Acknowledgments
The research was supported by Grants from the National Natural Science Foundation of China (Grant No. 81202101, 81172531, 61602119, and 30930038) and the Joint Technology Innovation Fund of Fujian Province (Grant number: 2016Y9044).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, H., Guo, W., Zhang, S. et al. A qualitative transcriptional signature to reclassify estrogen receptor status of breast cancer patients. Breast Cancer Res Treat 170, 271–277 (2018). https://doi.org/10.1007/s10549-018-4758-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-018-4758-2