Abstract
Purpose
LSD1 is overexpressed in various cancers including breast cancer, but its functional roles in tumourigenesis are not fully understood. This study aims at revealing the role of LSD1 in breast cancer development. In addition, it has been reported that phosphorylation of the Serine 112 residue of LSD1 by PKCα is crucial for its function in gene regulation. We also explored whether this phosphorylation affects LSD1’s role in breast cancer development.
Methods
This study includes LSD1 IHC data generated with tissue microarrays of 163 cases of breast cancer samples and 72 normal tissues. In vitro, role of LSD1, LSD1 S112D mutant (a phosphorylation simulation) and LSD1 S112A mutant (an unphosphorylation simulation) in induction of EMT is evaluated. Mechanismly, we checked the role of LSD1 and its mutant on E-cadherin promoter histone modifications. We also investigated the role of LSD1 and its mutants in metastasis with a nude mice model.
Results
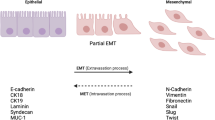
We found LSD1 is expressed at a higher level in breast cancer tissues compared with that in normal tissues, and LSD1 expression is closely linked to breast cancer metastasis. LSD1 potentiates EMT in breast epithelia cells by repressing E-cadherin expression through demethylating H3K4me at gene’s promoter, during which phosphorylation of LSD1 Ser112 is crucial for its binding and demethylation activity. In vivo, knockdown of LSD1 impairs the metastatic ability of MDA-MB-231 breast cancer cells in nude mice. Ectopic overexpression of either LSD1 or LSD1 S112D mutant (a phosphorylation simulation) facilitates metastasis, whereas the LSD1 S112A mutant (an unphosphorylation simulation) fails to affect the metastasis.
Conclusions
Data presented in this report indicate that LSD1 is able to induce EMT and to promote metastasis in breast cancer, and phosphorylation at LSD1 Ser112 is crucial for these functions.






Similar content being viewed by others
References
Bray F, McCarron P, Parkin DM (2004) The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res 6(6):229–239. doi:10.1186/bcr932
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. doi:10.1016/j.cell.2011.02.013
Gupta GP, Massague J (2006) Cancer metastasis: building a framework. Cell 127(4):679–695. doi:10.1016/j.cell.2006.11.001
Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA (2009) Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 119(6):1438–1449. doi:10.1172/JCI38019
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133(4):704–715. doi:10.1016/j.cell.2008.03.027
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139(5):871–890. doi:10.1016/j.cell.2009.11.007
Peinado H, Ballestar E, Esteller M, Cano A (2004) Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 24(1):306–319
Lin T, Ponn A, Hu X, Law BK, Lu J (2010) Requirement of the histone demethylase LSD1 in snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 29(35):4896–4904. doi:10.1038/onc.2010.234
Li Y, Wan X, Wei Y, Liu X, Lai W, Zhang L, Jin J, Wu C, Shao Q, Shao G, Lin Q (2016) LSD1-mediated epigenetic modification contributes to ovarian cancer cell migration and invasion. Oncol Rep 35(6):3586–3592. doi:10.3892/or.2016.4729
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119(7):941–953. doi:10.1016/j.cell.2004.12.012
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437(7057):436–439. doi:10.1038/nature04020
Lee MG, Wynder C, Cooch N, Shiekhattar R (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437(7057):432–435. doi:10.1038/nature04021
Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449(7158):105–108. doi:10.1038/nature06092
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41(1):125–129. doi:10.1038/ng.268
Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R (2011) Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer 128(3):574–586. doi:10.1002/ijc.25349
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y (2012) Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE 7(4):e35065. doi:10.1371/journal.pone.0035065
Wang M, Liu X, Guo J, Weng X, Jiang G, Wang Z, He L (2015) Inhibition of LSD1 by pargyline inhibited process of EMT and delayed progression of prostate cancer in vivo. Biochem Biophys Res Commun 467(2):310–315. doi:10.1016/j.bbrc.2015.09.164
Zhao ZK, Dong P, Gu J, Chen L, Zhuang M, Lu WJ, Wang DR, Liu YB (2013) Overexpression of LSD1 in hepatocellular carcinoma: a latent target for the diagnosis and therapy of hepatoma. Tumour Biol 34(1):173–180. doi:10.1007/s13277-012-0525-x
Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, Zhang Y, Yan ZS (2013) LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer 109(4):994–1003. doi:10.1038/bjc.2013.364
Serce N, Gnatzy A, Steiner S, Lorenzen H, Kirfel J, Buettner R (2012) Elevated expression of LSD1 (Lysine-specific demethylase 1) during tumour progression from pre-invasive to invasive ductal carcinoma of the breast. BMC Clin Pathol 12:13. doi:10.1186/1472-6890-12-13
Nagasawa S, Sedukhina AS, Nakagawa Y, Maeda I, Kubota M, Ohnuma S, Tsugawa K, Ohta T, Roche-Molina M, Bernal JA, Narvaez AJ, Jeyasekharan AD, Sato K (2015) LSD1 overexpression is associated with poor prognosis in basal-like breast cancer, and sensitivity to PARP inhibition. PLoS ONE 10(2):e0118002. doi:10.1371/journal.pone.0118002
Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J (2010) Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31(3):512–520. doi:10.1093/carcin/bgp324
Nam HJ, Boo K, Kim D, Han DH, Choe HK, Kim CR, Sun W, Kim H, Kim K, Lee H, Metzger E, Schuele R, Yoo SH, Takahashi JS, Cho S, Son GH, Baek SH (2014) Phosphorylation of LSD1 by PKCalpha is crucial for circadian rhythmicity and phase resetting. Mol Cell 53(5):791–805. doi:10.1016/j.molcel.2014.01.028
Metzger E, Willmann D, McMillan J, Forne I, Metzger P, Gerhardt S, Petroll K, von Maessenhausen A, Urban S, Schott AK, Espejo A, Eberlin A, Wohlwend D, Schule KM, Schleicher M, Perner S, Bedford MT, Jung M, Dengjel J, Flaig R, Imhof A, Einsle O, Schule R (2016) Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat Struct Mol Biol 23(2):132–139. doi:10.1038/nsmb.3153
Yang XJ, Seto E (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31(4):449–461. doi:10.1016/j.molcel.2008.07.002
Yang WH, Marth JD (2016) Protein glycosylation energizes T cells. Nat Immunol 17(6):613–614. doi:10.1038/ni.3468
Luo H, Shenoy AK, Li X, Jin Y, Jin L, Cai Q, Tang M, Liu Y, Chen H, Reisman D, Wu L, Seto E, Qiu Y, Dou Y, Casero RA Jr, Lu J (2016) MOF acetylates the histone demethylase LSD1 to suppress epithelial-to-mesenchymal transition. Cell Rep 15(12):2665–2678. doi:10.1016/j.celrep.2016.05.050
Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y (2009) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138(4):660–672. doi:10.1016/j.cell.2009.05.050
Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, Evers BM, Zhou BP (2013) The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell reports 5(1):224–236. doi:10.1016/j.celrep.2013.08.030
Bennani-Baiti IM (2012) Integration of ERalpha-PELP1-HER2 signaling by LSD1 (KDM1A/AOF2) offers combinatorial therapeutic opportunities to circumventing hormone resistance in breast cancer. Breast Cancer Res 14(5):112. doi:10.1186/bcr3249
Derr RS, van Hoesel AQ, Benard A, Goossens-Beumer IJ, Sajet A, Dekker-Ensink NG, de Kruijf EM, Bastiaannet E, Smit VT, van de Velde CJ, Kuppen PJ (2014) High nuclear expression levels of histone-modifying enzymes LSD1, HDAC2 and SIRT1 in tumor cells correlate with decreased survival and increased relapse in breast cancer patients. BMC Cancer 14:604. doi:10.1186/1471-2407-14-604
Ma L, Ma S, Zhao G, Yang L, Zhang P, Yi Q, Cheng S (2016) miR-708/LSD1 axis regulates the proliferation and invasion of breast cancer cells. Cancer Med 5(4):684–692. doi:10.1002/cam4.623
Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M (2006) Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci USA 103(38):13956–13961. doi:10.1073/pnas.0606381103
Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC (2011) LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 13(6):652–659. doi:10.1038/ncb2246
Venne AS, Kollipara L, Zahedi RP (2014) The next level of complexity: crosstalk of posttranslational modifications. Proteomics 14(4–5):513–524. doi:10.1002/pmic.201300344
Chen M, Qu X, Zhang Z, Wu H, Qin X, Li F, Liu Z, Tian L, Miao J, Shu W (2016) Cross-talk between Arg methylation and Ser phosphorylation modulates apoptosis signal-regulating kinase 1 activation in endothelial cells. Mol Biol Cell 27(8):1358–1366. doi:10.1091/mbc.E15-10-0738
Ying H, DePinho RA (2014) Cancer signaling: when phosphorylation meets methylation. Cell Res 24(11):1282–1283. doi:10.1038/cr.2014.103
McDermott SP, Wicha MS (2010) Targeting breast cancer stem cells. Mol Oncol 4(5):404–419. doi:10.1016/j.molonc.2010.06.005
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100(7):3983–3988. doi:10.1073/pnas.0530291100
Vianello P, Botrugno OA, Cappa A, Dal Zuffo R, Dessanti P, Mai A, Marrocco B, Mattevi A, Meroni G, Minucci S, Stazi G, Thaler F, Trifiro P, Valente S, Villa M, Varasi M, Mercurio C (2016) Discovery of a Novel Inhibitor of Histone Lysine-Specific Demethylase 1A (KDM1A/LSD1) as Orally Active Antitumor Agent. J Med Chem 59(4):1501–1517. doi:10.1021/acs.jmedchem.5b01209
Stazi G, Zwergel C, Valente S, Mai A (2016) LSD1 inhibitors: a patent review (2010-2015). Expert Opin Ther Pat 26(5):565–580. doi:10.1517/13543776.2016.1165209
Coover RA, Luzi NM, Korwar S, Casile ME, Lyons CE, Peterson DL, Ellis KC (2016) Design, synthesis, and in vitro evaluation of a fluorescently labeled irreversible inhibitor of the catalytic subunit of cAMP-dependent protein kinase (PKACalpha). Org Biomol Chem 14(20):4576–4581. doi:10.1039/c6ob00529b
Liang Q, Li L, Zhang J, Lei Y, Wang L, Liu DX, Feng J, Hou P, Yao R, Zhang Y, Huang B, Lu J (2013) CDK5 is essential for TGF-beta1-induced epithelial-mesenchymal transition and breast cancer progression. Sci Rep 3:2932. doi:10.1038/srep02932
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, Lu J, Huang B (2012) SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res 72(17):4597–4608. doi:10.1158/0008-5472.CAN-12-1045
Li L, Liu DX, Zhang N, Liang Q, Feng J, Yao M, Liu J, Li X, Zhang Y, Lu J, Huang B (2015) SHON, a novel secreted protein, regulates epithelial-mesenchymal transition through transforming growth factor-beta signaling in human breast cancer cells. Int J Cancer 136(6):1285–1295. doi:10.1002/ijc.29110
Nelson JD, Denisenko O, Bomsztyk K (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1(1):179–185. doi:10.1038/nprot.2006.27
Harmeyer KM, South PF, Bishop B, Ogas J, Briggs SD (2015) Immediate chromatin immunoprecipitation and on-bead quantitative PCR analysis: a versatile and rapid ChIP procedure. Nucleic Acids Res 43(6):e38. doi:10.1093/nar/gku1347
Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, Hou P, Gao Y, Zhao L, Wang G, Zhang Y, Liu DX, Huang B, Lu J (2014) PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res 74(19):5656–5667. doi:10.1158/0008-5472.CAN-14-0800
Acknowledgments
We thank Dr. Huang Wei from the School of Mathematics and Statistics of Northeast Normal University for kindly providing statistical analysis guidance. This work was supported by the grants from the National Natural Science Foundation of China (Grant Numbers 31570718, 31571478, 31571317, 31371294, 31271442 and 31401105) and from Jilin Scientific and Technological Development Program (Grant Numbers 20140520003JH and 20160101157JC). We also acknowledge the support from the “Program for Introducing Talents to Universities” (B07017) in assistance of international cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors disclose no potential conflicts of interest.
Authors’ contributions
J.F. contributed concept, designed and performed the experiments, analyzed the data and co-wrote the manuscript. GX, JL, NZ, LL and JJ participated in performing the experiments on breast cancer cell lines. LZ participated in IHC study. GW participated in athymismus mouse transplantation study. JZ, XW and JT made critical comments about the experiment designs. JL, BH and YZ supervised the project, participated in conception, design and data analysis and co-wrote the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Feng, J., Xu, G., Liu, J. et al. Phosphorylation of LSD1 at Ser112 is crucial for its function in induction of EMT and metastasis in breast cancer. Breast Cancer Res Treat 159, 443–456 (2016). https://doi.org/10.1007/s10549-016-3959-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3959-9