Abstract
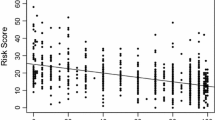
Molecular prognostic assays, such as Oncotype DX, are increasingly incorporated into the management of patients with invasive breast carcinoma. BreastPRS is a new molecular assay developed and validated from a meta-analysis of publically available genomic datasets. We applied the assay to matched fresh-frozen (FF) and formalin-fixed paraffin-embedded (FFPE) tumor samples to translate the assay to FFPE. A linear relationship of the BreastPRS prognostic score was observed between tissue preservation formats. BreastPRS recurrence scores were compared with Oncotype DX recurrence scores from 246 patients with invasive breast carcinoma and known Oncotype DX results. Using this series, a 120-gene Oncotype DX approximation algorithm was trained to predict Oncotype DX risk groups and then applied to series of untreated, node-negative, estrogen receptor (ER)-positive patients from previously published studies with known clinical outcomes. Correlation of recurrence score and risk group between Oncotype DX and BreastPRS was statistically significant (P < 0.0001). 59 of 260 (23 %) patients from four previously published studies were classified as intermediate-risk when the 120-gene Oncotype DX approximation algorithm was applied. BreastPRS reclassified the 59 patients into binary risk groups (high- vs. low-risk). 23 (39 %) patients were classified as low-risk and 36 (61 %) as high-risk (P = 0.029, HR: 3.64, 95 % CI: 1.40–9.50). At 10 years from diagnosis, the low-risk group had a 90 % recurrence-free survival (RFS) rate compared to 60 % for the high-risk group. BreastPRS recurrence score is comparable with Oncotype DX and can reclassify Oncotype DX intermediate-risk patients into two groups with significant differences in RFS. Further studies are needed to validate these findings.





Similar content being viewed by others
References
Van Laar RK (2011) Design and multiseries validation of a web-based gene expression assay for predicting breast cancer recurrence and patient survival. J Mol Diagn 13(3):297–304
Mittempergher L, de Ronde JJ, Nieuwland M et al (2011) Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS One 6:e17163
Passing H, Bablok (1983) A new biometrical procedure for testing the equality of measurements from two different analytical methods. Application of linear regression procedures for method comparison studies in clinical chemistry, part I. J Clin Chem Clin Biochem 21:709–720
Gentleman RC, Carey VJ, Bates DM et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
Dudoit S, Fridlyand J, Speed T (2002) Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc 97:77–87
Schmidt M, Bohm D, von Torne C et al (2008) The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res 68:5405–5413
Ivshina AV, George J, Senko O et al (2006) Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res 66:10292–10301
Loi S, Haibe-Kains B, Desmedt C et al (2007) Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 25:1239–1246
Desmedt C, Piette F, Loi S et al (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 13:3207–3214
Paik S, Tang G, Shak S et al (2006) Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24:3726–3734
Paik S, Shak S, Tang G et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Ma XJ, Patel R, Wang X et al (2006) Molecular classification of human cancers using a 92-gene real-time quantitative polymerase chain reaction assay. Arch Pathol Lab Med 130:465–473
Habel LA, Shak S, Jacobs MK et al (2006) A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res 8:R25
Albain KS, Barlow WE, Shak S et al (2010) Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol 11:55–65
Zujewski JA, Kamin L (2008) Trial assessing individualized options for treatment for breast cancer: the TAILORx trial. Future Oncol 4:603–610
Sparano JA (2006) TAILORx: trial assigning individualized options for treatment (Rx). Clin Breast Cancer 7:347–350
Disclosures
TMD declares no conflict of interest. RKVL is the Head of Bioinformatics and New Product Development for Signal Genetics and owns stock in the company. LTV declares no conflict of interest. WH, RF, NB, and LSJ are employees of Signal Genetics. SJS is a paid consultant of Signal Genetics.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
D’Alfonso, T.M., van Laar, R.K., Vahdat, L.T. et al. BreastPRS is a gene expression assay that stratifies intermediate-risk Oncotype DX patients into high- or low-risk for disease recurrence. Breast Cancer Res Treat 139, 705–715 (2013). https://doi.org/10.1007/s10549-013-2604-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2604-0