Abstract
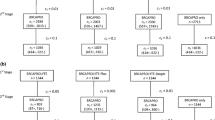
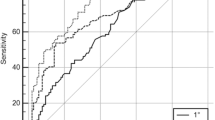
Health care providers need simple tools to identify patients at genetic risk of breast and ovarian cancers. Genetic risk prediction models such as BRCAPRO could fill this gap if incorporated into Electronic Medical Records or other Health Information Technology solutions. However, BRCAPRO requires potentially extensive information on the counselee and her family history. Thus, it may be useful to provide simplified version(s) of BRCAPRO for use in settings that do not require exhaustive genetic counseling. We explore four simplified versions of BRCAPRO, each using less complete information than the original model. BRCAPROLYTE uses information on affected relatives only up to second degree. It is in clinical use but has not been evaluated. BRCAPROLYTE-Plus extends BRCAPROLYTE by imputing the ages of unaffected relatives. BRCAPROLYTE-Simple reduces the data collection burden associated with BRCAPROLYTE and BRCAPROLYTE-Plus by not collecting the family structure. BRCAPRO-1Degree only uses first-degree affected relatives. We use data on 2,713 individuals from seven sites of the Cancer Genetics Network and MD Anderson Cancer Center to compare these simplified tools with the Family History Assessment Tool (FHAT) and BRCAPRO, with the latter serving as the benchmark. BRCAPROLYTE retains high discrimination; however, because it ignores information on unaffected relatives, it overestimates carrier probabilities. BRCAPROLYTE-Plus and BRCAPROLYTE-Simple provide better calibration than BRCAPROLYTE, so they have higher specificity for similar values of sensitivity. BRCAPROLYTE-Plus performs slightly better than BRCAPROLYTE-Simple. The Areas Under the ROC curve are 0.783 (BRCAPRO), 0.763 (BRCAPROLYTE), 0.772 (BRCAPROLYTE-Plus), 0.773 (BRCAPROLYTE-Simple), 0.728 (BRCAPRO-1Degree), and 0.745 (FHAT). The simpler versions, especially BRCAPROLYTE-Plus and BRCAPROLYTE-Simple, lead to only modest loss in overall discrimination compared to BRCAPRO in this dataset. Thus, we conclude that simplified implementations of BRCAPRO can be used for genetic risk prediction in settings where collection of complete pedigree information is impractical.



Similar content being viewed by others
References
Antoniou A, Pharoah PD, Narod S, et al (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72:1117–1130
King MC, Marks JH, Mandell JB (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302:643–646
Schwartz GF, Hughes KS, Lynch HT, Fabian CJ, Fentiman IS, Robson ME, Domchek SM, Hartmann LC, Holland R, Winchester DJ; Consensus Conference Committee The International Consensus Conference Committee. (2008) Proceedings of the international consensus conference on breast cancer risk, genetics, & risk management, April, 2007. Cancer 113:2627–2637
Drohan B, Roche CA, Cusack JC, Hughes KS (2012) Hereditary breast and ovarian cancer and other hereditary syndromes: using technology to identify carriers. Ann Surg Oncol 19:1732–1737
Drohan B, Ozanne EM, Hughes KS (2009) Electronic health records and the management of women at high risk of hereditary breast and ovarian cancer. Breast J 15(Suppl 1):46–55
Parmigiani G, Berry D, Aguilar O (1998) Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet 62:145–158
Chen S, Wang W, Broman KW, et al (2004) BayesMendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol 3:Article21
Tai YC, Chen S, Parmigiani G, et al (2008) Incorporating tumor immunohistochemical markers in BRCA1 and BRCA2 carrier prediction. Breast Cancer Res 10:401
Katki HA, Blackford A, Chen S, et al (2008) Multiple diseases in carrier probability estimation: accounting for surviving all cancers other than breast and ovary in BRCAPRO. Stat Med 27:4532–4548
Katki HA (2007) Incorporating medical interventions into Mendelian mutation prediction models. BMC Med Genet 8:13
Chen S, Blackford AL, Parmigiani G (2009) Tailoring BRCAPRO to Asian-Americans. J Clin Oncol 27:642–643
Biswas S, Tankhiwale N, Blackford A, et al (2012) Assessing the added value of breast tumor markers in genetic risk prediction model BRCAPRO. Breast Cancer Res Treat 133:347–355
Ozanne EM, Loberg A, Hughes S, et al (2009) Identification and management of women at high risk for hereditary breast/ovarian cancer syndrome. Breast J 15:155–162
Gilpin CA, Carson N, Hunter AG (2000) A preliminary validation of a family history assessment form to select women at risk for breast or ovarian cancer for referral to a genetics center. Clin Genet 58:299–308
Parmigiani G, Chen S, Iversen ES, et al (2007) Validity of models for predicting BRCA1 and BRCA2 mutations. Ann Intern Med 147:441–450
Chen S, Wang W, Lee S, et al (2006) Prediction of germline mutations and cancer risk in the Lynch syndrome. J Am Med Assoc 296:1479–1487
Pencina MJ, D’Agostino RB, D’Agostino RB, Vasan RS. (2008) Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 27:157–172
Efron B, Tibshirani R (1994) An introduction to the bootstrap. Chapman and Hall/CRC, Boca Raton
Parmigiani G (2002) Modeling in medical decision making: a Bayesian approach. Wiley, Chichester
Acknowledgments
This work was supported in part by Susan G Komen Grant KG081303, National Cancer Institute Grants 1R03CA173834-01 and 2P30CA006516-47 and the Dana Farber Cancer Institute.
Conflict of interest
The BayesMendel and HRA softwares are free to nonprofit institutions. HRA may be licensed to commercial entities, which would generate royalties for Hughes and Parmigiani.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Biswas, S., Atienza, P., Chipman, J. et al. Simplifying clinical use of the genetic risk prediction model BRCAPRO. Breast Cancer Res Treat 139, 571–579 (2013). https://doi.org/10.1007/s10549-013-2564-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2564-4