Abstract
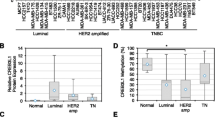
Klotho is a single pass transmembrane protein, associated with premature aging. We identified tumor suppressor activities for klotho, associated with reduced expression in breast cancer. We now aimed to analyze klotho expression in early stages of breast tumorigenesis and elucidate mechanisms leading to klotho silencing in breast tumors. We studied klotho expression, using immunohistochemistry, and found high klotho expression in all normal and mild hyperplasia samples, whereas reduced expression was associated with moderate and atypical ductal hyperplasia. Promoter methylation and histone deacetylation were studied as possible mechanisms for klotho silencing. Using bisulfite sequencing, and methylation-specific PCR, we identified KLOTHO promoter methylation in five breast cancer cell lines and in hyperplastic MCF-12A cells, but not in the non-tumorous mammary cell line HB2. Importantly, methylation status inversely correlated with klotho mRNA levels, and treatment of breast caner cells with 5-aza-2-deoxycytidine elevated klotho expression by up to 150-fold. KLOTHO promoter methylation was detected in 8/23 of breast cancer samples but not in normal breast samples. Chromatin immunoprecipitation revealed that in HB2 KLOTHO promoter was enriched with AcH3K9; however, in breast cancer cells, H3K9 was deacetylated, and treatment with the histone deacetylase inhibitor suberoylanilide bishydroxamide (SAHA) restored H3K9 acetylation. Taken together, these data indicate loss of klotho expression as an early event in breast cancer development, and suggest a role for DNA methylation and histone deacetylation in klotho silencing. Klotho expression and methylation may, therefore, serve as early markers for breast tumorigenesis.




Similar content being viewed by others
References
Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E et al (1997) Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390:45–51
Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H et al (2005) Suppression of aging in mice by the hormone Klotho. Science 309:1829–1833
Ito S, Kinoshita S, Shiraishi N, Nakagawa S, Sekine S, Fujimori T, Nabeshima Y (2000) Molecular cloning and expression analyses of mouse [beta]klotho, which encodes a novel Klotho family protein. Mech Develop 98:115–119
Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, Nabeshima Y (1998) Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun 242:626–630
Shiraki-Iida T, Aizawa H, Matsumura Y, Sekine S, Iida A, Anazawa H, Nagai R, Kuro-o M, Nabeshima Y (1998) Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein. FEBS Letters 424:6–10
Imura A, Iwano A, Tohyama O, Tsuji Y, Nozaki K, Hashimoto N, Fujimori T, Nabeshima Y (2004) Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Letters 565:143–147
Chen C-D, Podvin S, Gillespie E, Leeman SE, Abraham CR (2007) Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci 104:19796–19801
Ohyama Y, Kurabayashi M, Masuda H, Nakamura T, Aihara Y, Kaname T, Suga T, Arai M, Aizawa H, Matsumura Y et al (1998) Molecular cloning of rat klotho cDNA: markedly decreased expression of klotho by acute inflammatory stress. Biochem Biophys Res Commun 251:920–925
Cha S-K, Ortega B, Kurosu H, Rosenblatt KP, Kuro-o M, Huang C-L (2008) Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1. Proc Natl Acad Sci 105:9805–9810
Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu M-C, Moe OW et al (2006) Regulation of fibroblast growth factor-23 signaling by Klotho. J Biol Chem 281:6120–6123
Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T (2006) Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444:770–774
Wolf I, Sadetzki S, Catane R, Karasik A, Kaufman B (2005) Diabetes mellitus and breast cancer. Lancet Oncol 6:103–111
Yee D (2006) Targeting insulin-like growth factor pathways. Br J Cancer 94:465–468
Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, Kuro-o M, Karlan B, Kaufman B, Koeffler HP et al (2008) Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene 27:7094–7105
Wolf I, Laitman Y, Rubinek T, Abramovitz L, Novikov I, Beeri R, Kuro OM, Koeffler HP, Catane R, Freedman LS et al (2010) Functional variant of KLOTHO: a breast cancer risk modifier among BRCA1 mutation carriers of Ashkenazi origin. Oncogene 29:26–33
Chen B, Wang X, Zhao W, Wu J (2010) Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J Exp Clin Cancer Res 29:99
Doi S, Zou Y, Togao O, Pastor JV, John GB, Wang L, Shiizaki K, Gotschall R, Schiavi S, Yorioka N et al (2011) Klotho inhibits transforming growth factor-{beta}1 (TGF-{beta}1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J Biol Chem 286:8655–8665
Lee J, Jeong D-J, Kim J, Lee S, Park J-H, Chang B, Jung S-I, Yi L, Han Y, Yang Y et al (2010) The anti-aging gene KLOTHO is a novel target for epigenetic silencing in human cervical carcinoma. Molecular Cancer 9:109–119
Lu L, Katsaros D, Wiley A, de la Longrais IA, Puopolo M, Yu H (2008) Klotho expression in epithelial ovarian cancer and its association with insulin-like growth factors and disease progression. Cancer Invest 26:185–192
Wang X, Chen B, Xu W, Liu S, Zhao W, Wu J (2011) Combined effects of klotho and soluble CD40 ligand on A549 lung cancer cells. Oncol Rep 25(5):1465–1472
Sharma S, Kelly TK, Jones PA (2010) Epigenetics in cancer. Carcinogenesis 31:27–36
Lo P-K, Sukumar S (2008) Epigenomics and breast cancer. Pharmacogenomics 9:1879–1902
Wolf I, Bose S, Desmond J, Lin B, Williamson E, Karlan B, Koeffler H (2007) Unmasking of epigenetically silenced genes reveals DNA promoter methylation and reduced expression of PTCH in breast cancer. Breast Cancer Res Treat 105:139–155
Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP (2007) FOXA1: Growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer 120:1013–1022
McShane L, Altman D, Sauerbrei W, Taube S, Gion M, Clark G (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100:229–235
Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson Sm, Zhang X, Lin JC, Liang G, Jones PA, Tanay A (2008) Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci 105:12979–12984
Lacroix M, Leclercq G (2004) Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 83:249–289
Wolf I, O’Kelly J, Rubinek T, Tong M, Nguyen A, Lin BT, Tai H–H, Karlan BY, Koeffler HP (2006) 15-Hydroxyprostaglandin dehydrogenase is a tumor suppressor of human breast cancer. Cancer Res 66:7818–7823
Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA (1998) A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci 95:3003–3007
Schnitt SJ (2003) Benign breast disease and breast cancer risk: morphology and beyond. Am J Surg Pathol 27:836–841
Hoque MO, Prencipe M, Poeta ML, Barbano R, Valori VM, Copetti M, Gallo AP, Brait M, Maiello E, Apicella A et al (2009) Changes in CpG islands promoter methylation patterns during ductal breast carcinoma progression. Cancer Epidem Biomar 18:2694–2700
Herman JG, Merlo A, Mao L, Lapidus RG, Issa J-PJ, Davidson NE, Sidransky D, Baylin SB (1995) Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 55:4525–4530
Mancini DN, Rodenhiser DI, Ainsworth PJ, O’Malley FP, Singh SM, Xing W, Archer TK (1998) CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene 16:1161–1169
Schutte M, Rozenblum E, Moskaluk CA, Guan X, Hoque AT, Hahn SA, da Costa LT, de Jong PJ, Kern SE (1995) An integrated high-resolution physical map of the DPC/BRCA2 region at chromosome 13q12. Cancer Res 55:4570–4574
Geck P, Maffini MV, Szelei J, Sonnenschein C, Soto AM (2000) Androgen-induced proliferative quiescence in prostate cancer cells: the role of AS3 as its mediator. Proc Natl Acad Sci 97:10185–10190
Schott DR, Chang JN, Deng G, Kurisu W, Kuo WL, Gray J, Smith HS (1994) A candidate tumor suppressor gene in human breast cancers. Cancer Res 54:1393–1396
Acknowledgments
This research was supported by The Riva and Joel Koschitzky Fund for Breast Cancer Research at the Sheba Medical Center; the Chief Scientist Office of the Ministry of Health, Israel (grant no. 4055_3 to IW); the Israel Cancer Association Research Grant; The Israel Science Foundation (to I.W.); the “Talpiut” Sheba Career Development Award; and the Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
Conflicts of interest
The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rubinek, T., Shulman, M., Israeli, S. et al. Epigenetic silencing of the tumor suppressor klotho in human breast cancer. Breast Cancer Res Treat 133, 649–657 (2012). https://doi.org/10.1007/s10549-011-1824-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1824-4