Abstract
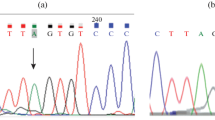
Germline mutations in BRCA1 were already linked to basal-like subtype of immunophenotypic molecular classification of breast cancer (BC). However, it is not known whether mutations in other BC susceptibility genes are associated with molecular subtypes of this cancer. We tested the hypothesis that distinct mutations in another BC susceptibility gene involved in DNA repair, i.e., CHEK2 may be associated with particular immunophenotypic molecular subtypes of this cancer. Two groups of patients: 1255 with BCs and 5496 healthy controls were genotyped for four CHEK2 mutations (I157T and three truncating mutations: 1100delC, IVS2 + 1G > A, del5395). BCs were tested by immunohistochemistry on tissue microarrays for ER, PR, HER-2, EGFR, and CK5/6 and were assigned to appropriate subtypes of immunophenotypic molecular classification. There was a significant association between CHEK2 mutations and the immunophenotypic molecular classification (P = 0.004). CHEK2-associated cancers were predominantly luminal (108/117 = 92.3%). CHEK2-I157T variant was associated with the luminal A subtype (P = 0.01), whereas CHEK2-truncating mutations were associated with the luminal B subtype (P = 0.005). Comparing the prevalence of CHEK2 mutations in BC with controls revealed that carriers of an I157T variant had OR of 1.80 for luminal A subtype and carriers of truncating mutations had OR of 6.26 for luminal B subtype of BC. To our knowledge, this is the first study showing that specific mutations in the same susceptibility gene are associated with different immunophenotypic molecular subtypes of BC. This association represents independent evidence supporting the biological significance of immunophenotypic molecular classification of BC.
Similar content being viewed by others
References
Antoni L, Sodha N, Collins I, Garrett MD (2007) CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat Rev Cancer 7:925–936
Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295:1379–1388
Sodha N, Mantoni TS, Tavtigian SV, Eeles R, Garrett MD (2006) Rare germ line CHEK2 variants identified in breast cancer families encode proteins that show impaired activation. Cancer Res 66:8966–8970
Wu X, Webster SR, Chen J (2001) Characterization of tumor-associated Chk2 mutations. J Biol Chem 276:2971–2974
Dong X, Wang L, Taniguchi K, Wang X, Cunningham JM, McDonnell SK, Qian C, Marks AF, Slager SL, Peterson BJ, Smith DI, Cheville JC, Blute ML, Jacobsen SJ, Schaid DJ, Tindall DJ, Thibodeau SN, Liu W (2003) Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet 72:270–280
Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H (2002) A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet 71:432–438
Falck J, Lukas C, Protopopova M, Lukas J, Selivanova G, Bartek J (2001) Functional impact of concomitant versus alternative defects in the Chk2–p53 tumour suppressor pathway. Oncogene 20:5503–5510
Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J (2001) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842–847
Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ (2002) Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 9:1045–1054
Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkila P, Aittomaki K, Holli K, Blomqvist C, Kallioniemi OP, Bartek J, Nevanlinna H (2004) CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer 111:543–547
Narod SA (2010) Testing for CHEK2 in the cancer genetics clinic: ready for prime time? Clin Genet 78:1–7
Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, Zlowocka E, Lenner M, Grabowska E, Nej K, Castaneda J, Medrek K, Szymanska A, Szymanska J, Kurzawski G, Suchy J, Oszurek O, Witek A, Narod SA, Lubinski J (2004) CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 75:1131–1135
Huzarski T, Cybulski C, Domagala W, Gronwald J, Byrski T, Szwiec M, Woyke S, Narod SA, Lubinski J (2005) Pathology of breast cancer in women with constitutional CHEK2 mutations. Breast Cancer Res Treat 90:187–189
Cybulski C, Gorski B, Huzarski T, Byrski T, Gronwald J, Debniak T, Wokolorczyk D, Jakubowska A, Kowalska E, Oszurek O, Narod SA, Lubinski J (2006) CHEK2-positive breast cancers in young Polish women. Clin Cancer Res 12:4832–4835
de Bock GH, Schutte M, Krol-Warmerdam EM, Seynaeve C, Blom J, Brekelmans CT, Meijers-Heijboer H, van Asperen CJ, Cornelisse CJ, Devilee P, Tollenaar RA, Klijn JG (2004) Tumour characteristics and prognosis of breast cancer patients carrying the germline CHEK2*1100delC variant. J Med Genet 41:731–735
Cybulski C, Huzarski T, Byrski T, Gronwald J, Debniak T, Jakubowska A, Gorski B, Wokolorczyk D, Masojc B, Narod SA, Lubinski J (2009) Estrogen receptor status in CHEK2-positive breast cancers: implications for chemoprevention. Clin Genet 75:72–78
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406:747–752
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98:10869–10874
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295:2492–2502
Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14:1368–1376
Tang P, Skinner KA, Hicks DG (2009) Molecular classification of breast carcinomas by immunohistochemical analysis: are we ready? Diagn Mol Pathol 18:125–132
Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkila P, Heikkinen T, Nevanlinna H, Akslen LA, Begin LR, Foulkes WD, Couch FJ, Wang X, Cafourek V, Olson JE, Baglietto L, Giles GG, Severi G, McLean CA, Southey MC, Rakha E, Green AR, Ellis IO, Sherman ME, Lissowska J, Anderson WF, Cox A, Cross SS, Reed MW, Provenzano E, Dawson SJ, Dunning AM, Humphreys M, Easton DF, Garcia-Closas M, Caldas C, Pharoah PD, Huntsman D (2010) Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 7:e1000279
Cybulski C, Wokolorczyk D, Huzarski T, Byrski T, Gronwald J, Gorski B, Debniak T, Masojc B, Jakubowska A, van de Wetering T, Narod SA, Lubinski J (2007) A deletion in CHEK2 of 5,395 bp predisposes to breast cancer in Poland. Breast Cancer Res Treat 102:119–122
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19:403–410
Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W (2011) Immunophenotypic predictive profiling of BRCA1-associated breast cancer. Virchows Arch 458:55–64
Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W (2011) PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy. Breast Cancer Res Treat 127:861–869
Chung GG, Kielhorn EP, Rimm DL (2002) Subjective differences in outcome are seen as a function of the immunohistochemical method used on a colorectal cancer tissue microarray. Clin Colorectal Cancer 1:237–242
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF, American Society of Clinical Oncology/College of American Pathologists (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med 131:18–43
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC (2010) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med 134:907–922
Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10:5367–5374
Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116:340–350
Durbecq V, Ameye L, Veys I, Paesmans M, Desmedt C, Sirtaine N, Sotiriou C, Bernard-Marty C, Nogaret JM, Piccart M, Larsimont D (2008) A significant proportion of elderly patients develop hormone-dependant “luminal-B” tumours associated with aggressive characteristics. Crit Rev Oncol Hematol 67:80–92
Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101:736–750
Bhargava R, Striebel J, Beriwal S, Flickinger JC, Onisko A, Ahrendt G, Dabbs DJ (2009) Prevalence, morphologic features and proliferation indices of breast carcinoma molecular classes using immunohistochemical surrogate markers. Int J Clin Exp Pathol 2:444–455
Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR (2010) Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol 4:255–266
Kilpivaara O, Bartkova J, Eerola H, Syrjakoski K, Vahteristo P, Lukas J, Blomqvist C, Holli K, Heikkila P, Sauter G, Kallioniemi OP, Bartek J, Nevanlinna H (2005) Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int J Cancer 113:575–580
Meyer A, Dork T, Sohn C, Karstens JH, Bremer M (2007) Breast cancer in patients carrying a germ-line CHEK2 mutation: Outcome after breast conserving surgery and adjuvant radiotherapy. Radiother Oncol 82:349–353
Schmidt MK, Tollenaar RA, de Kemp SR, Broeks A, Cornelisse CJ, Smit VT, Peterse JL, van Leeuwen FE, Van’t Veer LJ (2007) Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J Clin Oncol 25:64–69
Weischer M, Bojesen SE, Tybjaerg-Hansen A, Axelsson CK, Nordestgaard BG (2007) Increased risk of breast cancer associated with CHEK2*1100delC. J Clin Oncol 25:57–63
Chevallier B, Heintzmann F, Mosseri V, Dauce JP, Bastit P, Graic Y, Brunelle P, Basuyau JP, Comoz M, Asselain B (1988) Prognostic value of estrogen and progesterone receptors in operable breast cancer. Results of a univariate and multivariate analysis. Cancer 62:2517–2524
Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG (2008) CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol 26:542–548
Offit K, Garber JE (2008) Time to check CHEK2 in families with breast cancer? J Clin Oncol 26:519–520
Robson M (2010) CHEK2, breast cancer, and the understanding of clinical utility. Clin Genet 78:8–10
Acknowledgment
This study was supported by the Pomeranian Medical University Research Program grant #WL-125-01/S/11.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Domagala, P., Wokolorczyk, D., Cybulski, C. et al. Different CHEK2 germline mutations are associated with distinct immunophenotypic molecular subtypes of breast cancer. Breast Cancer Res Treat 132, 937–945 (2012). https://doi.org/10.1007/s10549-011-1635-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1635-7