Abstract
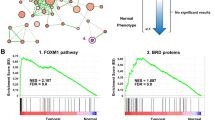
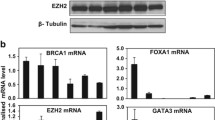
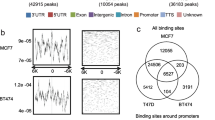
Expression profiling of BRCA1-deficient tumours has identified a pattern of gene expression similar to basal-like breast tumours. In this study, we examine whether a BRCA1-dependent transcriptional mechanism may underpin the link between BRCA1 and basal-like phenotype. In methods section, the mRNA and protein were harvested from a number of BRCA1 mutant and wild-type breast cancer cell lines and from matched isogenic controls. Microarray-based expression profiling was used to identify potential BRCA1-regulated transcripts. These gene targets were then validated (by in silico analysis of tumour samples) by real-time PCR and Western blot analysis. Chromatin immunoprecipitation (ChIP) assays were used to confirm recruitment of BRCA1 to specific promoters. In results, we demonstrate that functional BRCA1 represses the expression of cytokeratins 5(KRT5) and 17(KRT17) and p-Cadherin (CDH3) in HCC1937 and T47D breast cancer cell lines at both mRNA and protein level. ChIP assays demonstrate that BRCA1 is recruited to the promoters of KRT5, KRT17 and CDH3, and re-ChIP assays confirm that BRCA1 is recruited independently to form c-Myc and Sp1 complexes on the CDH3 promoter. We show that siRNA-mediated inhibition of endogenous c-Myc (and not Sp1) results in a marked increase in CDH3 expression analogous to that observed following the inhibition of endogenous BRCA1. The data provided suggest a model whereby BRCA1 and c-Myc form a repressor complex on the promoters of specific basal genes and represent a potential mechanism to explain the observed overexpression of key basal markers in BRCA1-deficient tumours.





Similar content being viewed by others
References
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406:747–752
Foulkes WD, Chappuis PO, Wong N, Brunet JS, Vesprini D, Rozen F, Yuan ZQ, Pollak MN, Kuperstein G, Narod SA, Begin LR (2000) Primary node negative breast cancer in BRCA1 mutation carriers has a poor outcome. Ann Oncol 11:307–313
Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA (2003) Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95:1482–1485
Foulkes WD, Brunet JS, Stefansson IM, Straume O, Chappuis PO, Begin LR, Hamel N, Goffin JR, Wong N, Trudel M, Kapusta L, Porter P, Akslen LA (2004) The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res 64:830–835
Arnes JB, Brunet JS, Stefansson I, Begin LR, Wong N, Chappuis PO, Akslen LA, Foulkes WD (2005) Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 11:4003–4011
Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, Rodriguez S, Cigudosa JC, Diez O, Alonso C, Lerma E, Dopazo J, Rivas C, Benitez J (2005) Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res Treat 90:5–14
Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, Farid LM, Venter D, Antoniou A, Storfer-Isser A, Smyth E, Steel CM, Haites N, Scott RJ, Goldgar D, Neuhausen S, Daly PA, Ormiston W, McManus R, Scherneck S, Ponder BA, Ford D, Peto J, Stoppa-Lyonnet D, Easton DF et al (1998) Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst 90:1138–1145
Lakhani SR, O’Hare MJ, Ashworth A (2001) Profiling familial breast cancer. Nat Med 7:408–410
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100:8418–8423
Grushko TA, Dignam JJ, Das S, Blackwood AM, Perou CM, Ridderstrale KK, Anderson KN, Wei MJ, Adams AJ, Hagos FG, Sveen L, Lynch HT, Weber BL, Olopade OI (2004) MYC is amplified in BRCA1-associated breast cancers. Clin Cancer Res 10:499–507
Li H, Lee TH, Avraham H (2002) A novel tricomplex of BRCA1, Nmi, and c-Myc inhibits c-Myc-induced human telomerase reverse transcriptase gene (hTERT) promoter activity in breast cancer. J Biol Chem 277:20965–20973
Li Y, Pan J, Li JL, Lee JH, Tunkey C, Saraf K, Garbe JC, Whitley MZ, Jelinsky SA, Stampfer MR, Haney SA (2007) Transcriptional changes associated with breast cancer occur as normal human mammary epithelial cells overcome senescence barriers and become immortalized. Mol Cancer 6:7
Kennedy RD, Gorski JJ, Quinn JE, Stewart GE, James CR, Moore S, Mulligan K, Emberley ED, Lioe TF, Morrison PJ, Mullan PB, Reid G, Johnston PG, Watson PH, Harkin DP (2005) BRCA1 and c-Myc associate to transcriptionally repress psoriasin, a DNA damage-inducible gene. Cancer Res 65:10265–10272
Hosey AM, Gorski JJ, Murray MM, Quinn JE, Chung WY, Stewart GE, James CR, Farragher SM, Mulligan JM, Scott AN, Dervan PA, Johnston PG, Couch FJ, Daly PA, Kay E, McCann A, Mullan PB, Harkin DP (2007) Molecular basis for estrogen receptor alpha deficiency in BRCA1-linked breast cancer. J Natl Cancer Inst 99:1683–1694
Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE (2006) Rosetta error model for gene expression analysis. Bioinformatics 22:1111–1121
Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, Harkin DP (2003) BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res 63:6221–6228
Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, Merajver SD, Ethier SP, Schutte M (2006) BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res 66:41–45
Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F, Evans G, Maugard C, Meijers-Heijboer H, Klijn JG, Olah E, Gusterson BA, Pilotti S, Radice P, Scherneck S, Sobol H, Jacquemier J, Wagner T, Peto J, Stratton MR, McGuffog L, Easton DF (2005) Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 11:5175–5180
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527
Mullan PB, Quinn JE, Harkin DP (2006) The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25:5854–5863
Laakso M, Loman N, Borg A, Isola J (2005) Cytokeratin 5/14-positive breast cancer: true basal phenotype confined to BRCA1 tumors. Mod Pathol 18:1321–1328
Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, Tutt AN (2007) BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26:2126–2132
Abramovitch S, Glaser T, Ouchi T, Werner H (2003) BRCA1-Sp1 interactions in transcriptional regulation of the IGF-IR gene. FEBS Lett 541:149–154
Wang Q, Zhang H, Kajino K, Greene MI (1998) BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene 17:1939–1948
Palacios J, Robles-Frias MJ, Castilla MA, Lopez-Garcia MA, Benitez J (2008) The molecular pathology of hereditary breast cancer. Pathobiology 75:85–94
Author information
Authors and Affiliations
Corresponding author
Additional information
Funding bodies
Julia Gorski and Niamh Buckley: R&D Office NI, Colin James: Cancer Research UK, Jennifer Quinn: Breast Cancer Campaign, Gail Stewart: Medical Research Council, Kieran Crosbie Staunton: European Social Fund, Richard Wilson: Queens University Belfast, Richard Kennedy and Fionnuala McDyer: Almac Diagnostics, Paul Mullan: Action Cancer N. I., Paul Harkin: Breast Cancer Campaign and Cancer Research UK.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gorski, J.J., James, C.R., Quinn, J.E. et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res Treat 122, 721–731 (2010). https://doi.org/10.1007/s10549-009-0565-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-009-0565-0