Abstract
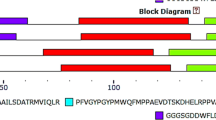
ZAT12 a C2H2-zinc-finger protein is an abiotic stress-responsive transcription factor in plants having less information about their structure. Transcription analysis proved that ZAT12 transcripts over-expressed during drought, heat and salt stress conditions which led to an interest in 3-D structural studies of ZAT12in Brassica carinata. Over-expression of BcZAT12 in transformed tomato plants under abiotic stresses, suggest role of ZAT12 in conferring stress-tolerance in tomato. Sequence analysis of ZAT12 protein (Accession No. ABB55254.1) from B. carinata revealed it as a 161 amino acid long protein with short conserved motif 140LDLXL144 in C-terminal, a leucine rich L-Box with—14EXXAXCLXXL23 motif in N-terminal region and presence of two conserved Zinc-Finger motifs “CXXCXXXXXXXQALGGHXXXH” between positions 42–62 and 85–105. The two zinc finger motifs have presence of two conserved glutamic acid (Glu) and phenylalanine (Phe) residues. Two methionine (Met) residues at position 94 and 102 present in ZF-motif-2 were absent in ZF-motif-1. The 94Met and 97Ala in ZF-motif-2 were found to be replaced by serine (Ser) in ZF-motif-1. Homology and ab initio structural modeling of ZAT12 encoded BcZAT12 protein of B. carinata resulted in robust 3-D models and were evaluated for structural motifs, associated GO terms and protein-DNA interactions. The BcZAT12 protein model, was of good quality, reliable, stable and is deposited in PMDB database (PMDB ID: PM0078213). BcZAT12 is annotated as an intracellular protein having molecular function in Zn-binding which in turn regulates signal transduction/translation processes in response to abiotic stresses in plants. Results suggest BcZAT12 protein to interact directly with one strand of dsDNA via electrostatic and H-bonds.












Similar content being viewed by others
References
Balla S, Thapar V, Verma S, Luong T, Faghri T et al (2006) Minimotif Miner: a tool for investigating protein function. Nat Methods 3:175–177
Baltz R, Domon C, Pillay DTN, Steinmetz A (1992) Characterisation of a pollen-specific cDNA from sunflower encoding a zinc finger protein. Plant J 2:713–721
Bernstein FC, Koetzle TF, Williams GJB, Meyer EF Jr, Brice MD et al (1997) The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol 112:535–542
Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC (eds) Bioinformatics for Geneticists (hierarchical exotoxicology mini series) Chichester. Wiley, UK, pp 289–316
Bowie JU, Luthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170
Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282:9260–9268
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of non bonded atomic interactions. Protein Sci 2:1511–1519
Davletova S, Schlauch K, Coutu J, Mittler R (2005) The Zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139:847–856
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 8:2714–2723
Kiełbowicz-Matuk A (2012) Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci 185–186:78–85
Laskowski RA, McArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Li Y, Zhang Y (2009) REMO: a new protocol to refine full atomic protein models from C-α traces by optimizing hydrogen-bonding networks. Proteins 76:665–676
Livaka KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Lu D, Klug A (2007) Invariance of the zinc finger module: a comparison of the free structure with those in nucleic-acid complexes. Proteins 67:508–512
Meissner R, Michael AJ (1997) Isolation and characterisation of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol Biol 33:615–624
Miller J, McLachlan AD, Klug A (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4:1609–1614
Nahakpam S, Shah K (2011) Expression of key antioxidant enzymes under combined effect of heat and cadmium toxicity in growing rice seedlings. Plant Growth Regul 63:23–35
Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J et al (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104:16450–16455
Rai AC, Singh M, Shah K (2012a) Environmental stresses and transgenics: Role of ZFP (ZAT) gene in multiple stress tolerance in plants. In: Hemantranjan A (ed) Advances in plant physiology. Scientific Publishers, Jodhpur, India, pp 197–232
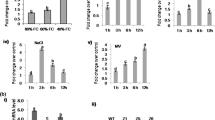
Rai AC, Singh M, Shah K (2012b) Effect of water withdrawal on formation of free radical, proline accumulation and activities of antioxidant enzymes in ZAT12-transformed transgenic tomato plants. Plant Physiol Biochem 61:108–114
Rai AC, Singh M, Shah K (2013) Engineering drought tolerant tomato plants over-expressing BcZAT12 gene encoding a C2H2 zinc finger transcription factor. Phytochemistry 85:44–50
Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279:11736–11743
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738
Sakamoto H, Araki T, Meshi T, Iwabuchi M (2000) Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress. Gene 248:23–32
Schauser L, Christensen L, Borg S, Poulsen C (1995) PZF, a cDNA isolated from Lotus japonicus and soybean root nodule libraries, encodes a new plant member of the RING-finger family of zinc-binding proteins. Plant Physiol 107:1457–1458
Shah K, Singh M, Rai AC (2013) Effect of heat-shock induced oxidative stress is suppressed in BcZAT12 expressing drought tolerant tomato. Phytochemistry 95:109–117
Singh I, Shah K (2012) In silico study of interaction between rice proteins enhanced disease susceptibility 1 and phytoalexin deficient 4, the regulators of salicylic acid signalling pathway. J Biosci 237:563–571
Singh I, Agrawal P, Shah K (2012) In search of function for hypothetical proteins encoded by genes of SA-JA pathways in Oryza sativa by in silico comparison and structural modeling. Bioinformation 8:001–005
Sun S-J, Guo S-Q, Yang X, Bao Y-M, Tang H-J et al (2010) Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot 61:2807–2818
Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R (2011) Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J 66:844–851
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ul-Haq Z, Khan W, Zarina S, Sattar R, Moin ST (2010) Template-based structure prediction and molecular dynamics simulation study of two mammalian Aspartyl-tRNA synthetases. J Mol Graph Modell 28:401–412
Wu S, Skolnick J, Zhang Y (2007) Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol 5:17
Zhang Y (2007) Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 69:118–128
Zhang Y (2009a) Protein structure prediction: when is it useful? Curr Opin Struct Biol 19:145–155
Zhang Y (2009b) I-TASSER: fully automated protein structure prediction in CASP8. Proteins 77:100–103
Acknowledgments
The work is a part of Network Project on Transgenic Crops, funded by ICAR, New Delhi. The authors are grateful to the Bioinformatics Division, MMV, BHU for computational facilities and to www.molegro.com/mvd-products.php server for access to Molegro Virtual docker for conducting the research work. All computations were performed on linux high performance clusters. Authors are grateful to Mr. Gerard Charmasson for copyediting of manuscript for language usage, spelling and grammar.
Conflict of interest
The authors declare that there are no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rai, A.C., Singh, I., Singh, M. et al. Expression of ZAT12 transcripts in transgenic tomato under various abiotic stresses and modeling of ZAT12 protein in silico. Biometals 27, 1231–1247 (2014). https://doi.org/10.1007/s10534-014-9785-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-014-9785-9