Abstract
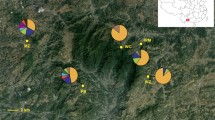
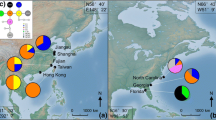
Ambrosia artemisiifolia is native to North America but has become a worldwide invasive weed. It was introduced to China more than 80 years ago and has spread into 20 provinces since then. To assess the population structure of A. artemisiifolia in China and whether this invasion involved a single event or multiple events, we investigated patterns of genetic variation for three chloroplast DNA intergenic spacer regions, a nrITS region and five microsatellite loci. Our dataset consists of 370 individuals from 19 sites throughout China. We compared their cpDNA-haplotypes to those published for native North American populations. The distribution of cpDNA-haplotypes indicates that A. artemisiifolia was introduced to China multiple times from different source regions. The numbers of alleles in Chinese populations were not significantly lower than in native populations. Both nrITS-haplotypes and microsatellite alleles showed that there was no evidence for a genetic bottleneck. Four populations were genetically well separated from the other 15 populations. However, the absence of isolation by distance, and the low levels of genetic differentiation and gene flow among the other 15 population suggest that most populations in China come from pre-admixed populations. To find the exact source regions of the Chinese populations, more samples from the native region and other invaded regions will be necessary. Nevertheless, our study provides important insights into the genetic background of A. artemisiifolia invasion in China.






Similar content being viewed by others
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Berbegal M, Armengol J, Grünwald NJ (2013) Evidence for multiple introductions and clonality in spanish populations of Fusarium circinatum. Phytopathology 103:851–861
Blossey B, Notzold R (1995) Evolution of increased in invasive competitive ability nonindigenous a hypothesis plants: a hypothesis. J Ecol 83:887–889
Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D (2005) Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144:1–11
Cao LJ, Wang ZH, Gong YJ, Zhu L, Hoffmann AA, Wei SJ (2017) Low genetic diversity but strong population structure reflects multiple introductions of western flower thrips (Thysanoptera: Thripidae) into China followed by human-mediated spread. Evol Appl 10:391–401
Chun YJ, Fumanal B, Laitung B, Bretagnolle F (2010) Gene flow and population admixture as the primary post-invasion processes in common ragweed (Ambrosia artemisiifolia) populations in France. New Phytol 185:1100–1107
Chun YJ, Le Corre V, Bretagnolle F (2011) Adaptive divergence for a fitness-related trait among invasive Ambrosia artemisiifolia populations in France. Mol Ecol 20:1378–1388
Ciappetta S, Ghiani A, Gilardelli F, Bonini M, Citterio S, Gentili R (2016) Invasion of Ambrosia artemisiifolia in Italy: assessment via analysis of genetic variability and herbarium data. Flora 223:106–113
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Di Rienzo A, Peterson AC, Garzat JC, Valdes AM, Slatkint M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Dlugosch KM, Anderson SR, Braasch J, Cang FA, Gillette HD (2015) The devil is in the details: genetic variation in introduced populations and its contributions to invasion. Mol Ecol 24:2095–2111
Dong H, Zhou M, Liu Z, Hao X, Liu Y, Abdulvai A, Liu T (2017) Diffusion and intrusion features of Ambrosia artemisiifolia and Ambrosia trifida in Yili River Valley. J Arid Land Res Enviorn 11:175–180
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellstrand NC, Schierenbeck KA (2006) Hybridization as a stimulus for the evolution of invasiveness in plants? Euphytica 148:35–46
Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B (2016) Is there a genetic paradox of biological invasion? Annu Rev Ecol Evol Syst 47:51–72
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2006) ARLEQUIN version 3.1: a software for population genetic data analysis. Computational and molecular population genetics laboratory. Institute of Zoology, University of Berne, Bern
Fischer ML, Salgado I, Beninde J et al (2017) Multiple founder effects are followed by range expansion and admixture during the invasion process of the raccoon (Procyon lotor) in Europe. Divers Distrib 23:409–420
Flot JF (2010) SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol Ecol Resour 10:162–166
Friedman J, Barrett SCH (2008) High outcrossing in the annual colonizing species Ambrosia artemisiifolia (Asteraceae). Ann Bot 101:1303–1309
Gaudeul M, Giraud T, Kiss L, Shykoff JA (2011) Nuclear and chloroplast microsatellites show multiple introductions in the worldwide invasion history of common ragweed, Ambrosia artemisiifolia. PLoS ONE 6:e17658
Gentili R, Montagnani C, Gilardelli F, Guarino MF, Citterio S (2017) Let native species take their course: Ambrosia artemisiifolia replacement during natural or “artificial” succession. Acta Oecol 82:32–40
Genton BJ, Shykoff JA, Giraud T (2005) High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol Ecol 14:4275–4285
Gladieux P, Giraud T, Kiss L, Genton BJ, Jonot O, Shykoff JA (2011) Distinct invasion sources of common ragweed (Ambrosia artemisiifolia) in Eastern and Western Europe. Biol Invasions 13:933–944
Groves RH, Burdon JJ (1986) Ecology of biological invasions: an Australian perspective. Australian Academy of Science, Canberra
Guillot G, Santos F, Estoup A (2008) Analysing georeferenced population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 24:1406–1407
Harrigan RJ, Mazza ME, Sorenson MD (2008) Computation vs cloning: evaluation of two methods for haplotype determination. Mol Ecol Resour 8:1239–1248
Henry P, Le Lay G, Goudet J, Guisan A, Jahodová S, Besnard G (2009) Reduced genetic diversity, increased isolation and multiple introductions of invasive giant hogweed in the western Swiss Alps. Mol Ecol 18:2819–2831
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jin DP, Lee JH, Xu B, Choi BH (2016) Phylogeography of East Asian Lespedeza buergeri (Fabaceae) based on chloroplast and nuclear ribosomal DNA sequence variations. J Plant Res 129:793–805
Karn E, Jasieniuk M (2017) Genetic diversity and structure of Lolium perenne ssp. multiflorum in California vineyards and orchards indicate potential for spread of herbicide resistance via gene flow. Evol Appl 10:616–629
Keller SR, Taylor DR (2010) Genomic admixture increases fitness during a biological invasion. J Evol Biol 23:1720–1731
Keller M, Kollmann J, Edwards PJ (2000) Genetic introgression from distant provenances reduces fitness in local weed populations. J Appl Ecol 37:647–659
Keller SR, Fields PD, Berardi AE, Taylor DR (2014) Recent admixture generates heterozygosity-fitness correlations during the range expansion of an invading species. J Evol Biol 27:616–627
Kropf M, Huppenberger AS, Karrer G (2018) Genetic structuring and diversity patterns along rivers—local invasion history of Ambrosia artemisiifolia (Asteraceae) along the Danube River in Vienna (Austria) shows non-linear pattern. Weed Res 58:131–140
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Ecol Resour 33:1870–1874
Lawalrée A (1955) Note complémentaire sur les Ambrosia adventices en Europe occidentale. Bull Soc R Bot Belg 87:207–208
Leberg PL (1992) Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution 46:477–494
Li XM, Liao WJ, Wolfe LM, Zhang DY (2012) No evolutionary shift in the mating system of North American Ambrosia artemisiifolia (Aasteraceae) following its introduction to china. PLoS ONE 7:1–6
Li XM, Zhang DY, Liao WJ (2015) The rhythmic expression of genes controlling flowering time in southern and northern populations of invasive Ambrosia artemisiifolia. J Plant Ecol 8:207–212
Li Y, Stift M, Kleunen M (2018) Admixture increases performance of an invasive plant beyond first generation heterosis. J Ecol 106:1595–1606
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu J, Chen H, Kowarik I, Zhang Y, Wang R (2012) Plant invasions in China: an emerging hot topic in invasion science. NeoBiota 15:27–51
Lockwood JL, Hoopes MF, Marchetti MP (2007) Invasion ecology. Wiley, Oxford
Makra L, Matyasovszky I, Hufnagel L, Tusnady G (2015) The history of ragweed in the world. Appl Ecol Env Res 13:489–512
Martin MD, Chamecki M, Brush GS, Meneveau C, Parlange MB (2009) Pollen clumping and wind dispersal in an invasive angiosperm. Am J Bot 96:1703–1711
Martin MD, Zimmer EA, Olsen MT, Foote AD, Gilbert MTP, Brush GS (2014) Herbarium specimens reveal a historical shift in phylogeographic structure of common ragweed during native range disturbance. Mol Ecol 23:1701–1716
Martin MD, Olsen MT, Samaniego JA, Zimmer EA, Gilbert MTP (2016) The population genomic basis of geographic differentiation in North American common ragweed (Ambrosia artemisiifolia L.). Ecol Evol 6:3760–3771
Meyer L, Causse R, Pernin F et al (2017) New gSSR and EST-SSR markers reveal high genetic diversity in the invasive plant Ambrosia artemisiifolia L. and can be transferred to other invasive Ambrosia species. PLoS ONE 12:97
Molecular Ecology Resources Primer Development Consortium (MERPDC) (2009) Permanent genetic resources added to Molecular Ecology Resources database 1 January 2009–30 April 2009. Mol Ecol Resour 9:1375–1429
Montagnani C, Gentili R, Smith M, Guarino MF, Citterio S (2017) The worldwide spread, success, and impact of ragweed (Ambrosia spp.). Crit Rev Plant Sci 36:139–178
Müller-Schärer H, Schaffner U, Steinger T (2004) Evolution in invasive plants: implications for biological control. Trends Ecol Evol 19:417–422
Naegele RP, Tomlinson AJ, Hausbeck MK (2015) Evaluation of a diverse, worldwide collection of wild, cultivated, and landrace pepper (Capsicum annuum) for resistance to phytophthora fruit rot, genetic diversity, and population structure. Phytopathology 105:110–118
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Pagad S, Genovesi P, Carnevali L, Schigel D, McGeoch MA (2018) Data descriptor: introducing the global register of introduced and invasive species. Sci Data 5:170202
Pairon M, Petitpierre B, Campbell M et al (2010) Multiple introductions boosted genetic diversity in the invasive range of black cherry (Prunus serotina; Rosaceae). Ann Bot 105:881–890
Parks DH, Mankowski T, Zangooei S et al (2013) GenGIS 2: geospatial analysis of traditional and genetic biodiversity, with new gradient algorithms and an extensible plugin framework. PLoS ONE 8:e69885
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel population genetic software for teaching and research. Mol Ecol Resour 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 65: genetic analysis in excel population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pointier J, Jarne P, Sarda V, David P, Spe I (2008) Report high genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr Biol 18:363–367
Polzin T, Daneshmand SV (2003) On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett 31:12–20
Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ (2008) Adaptive evolution in invasive species. Trends Plant Sci 13:288–294
Priszter S (1960) Adventív gyomnövényeink terjedése. Mezögazdasági Kiadó Agricultural Publishing Ltd, Budapest
Pritchard J, Wen X, Falush D (2010) Documentation for STRUCTURE software, version 23. University of Chicago, Chicago
Pyšek P, Pergl J, Essl F et al (2017) Naturalized alien flora of the world: species diversity, taxonomic and phylogenetic patterns, geographic distribution and global hotspots of plant invasion. Preslia 89:203–274
Rius M, Darling JA (2014) How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol 29:233–242
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:454–464
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Resour 4:137–138
Schierenbeck KA, Ellstrand NC (2009) Hybridization and the evolution of invasiveness in plants and other organisms. Biol Invasions 11:1093–1105
Shi J, Macel M, Tielbörger K, Verhoeven KJ (2018) Effects of admixture in native and invasive populations of Lythrum salicaria. Biol Invasions 20:2381–2393
Signorile AL, Lurz PWW, Wang J, Reuman DC, Carbone C (2016) Mixture or mosaic? Genetic patterns in UK grey squirrels support a human-mediated “long-jump” invasion mechanism. Divers Distrib 22:566–577
Stephens M, Donnelly P (2003) A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
The People’s Government of Meizhou (2016) Meizhou overview. https://www.meizhou.gov.cn/zjmz/mzgk/mzgk1/t20160825_130.htm. Accessed 12 Dec 2018
Tsutsui K, Suwa A, Sawada KI, Kato T, Ohsawa TA, Watano Y (2009) Incongruence among mitochondrial, chloroplast and nuclear gene trees in Pinus subgenus Strobus (Pinaceae). J Plant Res 122:509–521
van Boheemen LA, Lombaert E, Nurkowski KA et al (2017) Multiple introductions, admixture and bridgehead invasion characterize the introduction history of Ambrosia artemisiifolia in Europe and Australia. Mol Ecol 26:5421–5434
van Kleunen M, Bossdorf O, Dawson W (2018) The ecology and evolution of alien plants. Annu Rev Ecol Evol Syst 49:25–47
Verhoeven KJ, Macel M, Wolfe LM, Biere A (2010) Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc R Soc B 278:2–8
Wan FH, Guo JY, Zhang F (2009) Research on biological invasions in China. Science Press, Beijing
Wan F, Jiang M, Zhan A (2017) Biological invasions and its management in china, vol 2. Springer, Singapore
Wang MY (2005) Current situation of common ragweed and control strategy. J Anhui Agric Sci 33:1771–1786
Wang J, Wu Y, Ren G, Guo Q, Liu J, Lascoux M (2011) Genetic differentiation and delimitation between ecologically diverged Populus euphratica and P. pruinosa. PLoS ONE 6:e26530
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Zhou ZS, Guo JY, Wan FH (2015) Ambrosia artemisiifolia L. In: Wan FH, Hou YM, Jiang MX (eds) Invasion biology. Science Press, Beijing, pp 182–185
Acknowledgements
The authors would like to thank Dr. Michael D. Martin at the Centre for GeoGenetics, Natural History Museum of Denmark for providing haplotypes sequences from the 45 North American extant populations and historical Ambrosia artemisiifolia herbarium specimen.
Funding
This work was financially supported by grants from The National Key Research and Development Program of China 2016YFC1201100, 2017YFC0506200, and The National Natural Science Foundation of China 41701026.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, F., van Kleunen, M., Li, J. et al. Patterns of genetic variation reflect multiple introductions and pre-admixture sources of common ragweed (Ambrosia artemisiifolia) in China. Biol Invasions 21, 2191–2209 (2019). https://doi.org/10.1007/s10530-019-01966-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-019-01966-2