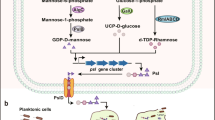
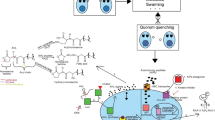
Abstract
Cell–cell communication in bacteria needs chemical signals and cognate receptors. Many Gram-negative bacteria use acyl-homoserine lactones (AHLs) and cognate LuxR-type receptors to regulate their quorum sensing (QS) systems. The signal synthase-receptor (LuxI–LuxR) pairs may have co-evolved together. However, many LuxR solo (orphan LuxR) regulators sense more signals than just AHLs, and expand the regulatory networks for inter-species and inter-kingdom communication. Moreover, there are also some QS regulators from the TetR family. LuxR solo regulators might have evolved by gene duplication and horizontal gene transfer. An increased understanding of the evolutionary roles of QS regulators would be helpful for engineering of cell–cell communication circuits in bacteria.


Similar content being viewed by others
References
Alguel Y, Meng C, Teran W, Krell T, Ramos JL, Gallegos MT, Zhang X (2007) Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J Mol Biol 369:829–840
Amin SA et al (2015) Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522:98
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–W350
Ball AS, Chaparian RR, van Kessel JC (2017) Quorum sensing gene regulation by LuxR/HapR master regulators in Vibrios. J Bacteriol 199:e00105–e00117
Biarnes-Carrera M, Breitling R, Takano E (2015) Butyrolactone signalling circuits for synthetic biology. Curr Opin Chem Biol 28:91–98
Biasini M et al (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258
Bottomley MJ, Muraglia E, Bazzo R, Carfi A (2007) Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem 282:13592–13600
Brachmann AO et al (2013) Pyrones as bacterial signaling molecules. Nat Chem Biol 9:573–573
Brameyer S, Heermann R (2017) Quorum sensing and LuxR solos in Photorhabdus. Curr Top Microbiol Immunol 402:103–119
Brameyer S, Kresovic D, Bode HB, Heermann R (2015) Dialkylresorcinols as bacterial signaling molecules. Proc Natl Acad Sci USA 112:572–577
Camilli A, Bassler BL (2006) Bacterial small-molecule signaling pathways. Science 311:1113–1116
Chen G et al (2011) A strategy for antagonizing quorum sensing. Mol Cell 42:199–209
Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549
Chugani S, Greenberg EP (2014) An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR. Front Cell Infect Microbiol 4:152
Coutinho BG, Mevers E, Schaefer AL, Pelletier DA, Harwood CS, Clardy J, Greenberg EP (2018) A plant-responsive bacterial-signaling system senses an ethanolamine derivative. Proc Natl Acad Sci USA 115:9785–9790
Cuthbertson L, Nodwell JR (2013) The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475
Daniel R, Rubens JR, Sarpeshkar R, Lu TK (2013) Synthetic analog computation in living cells. Nature 497:619–623
De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ (2007) Crystal structure of the Vibrio cholerae quorum-sensing regulatory protein HapR. J Bacteriol 189:5683–5691
Frenois F, Engohang-Ndong J, Locht C, Baulard AR, Villeret V (2004) Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol Cell 16:301–307
Gonzalez JF, Myers MP, Venturi V (2013) The inter-kingdom solo OryR regulator of Xanthomonas oryzae is important for motility. Mol Plant Pathol 14:211–221
Gonzalez JF, Venturi V (2013) A novel widespread interkingdom signaling circuit. Trends Plant Sci 18:167–174
Gray KM, Garey JR (2001) The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379–2387
Ha C, Park SJ, Im SJ, Park SJ, Lee JH (2012) Interspecies signaling through QscR, a quorum receptor of Pseudomonas aeruginosa. Mol Cells 33:53–59
Hao G, Burr TJ (2006) Regulation of long-chain N-acyl-homoserine lactones in Agrobacterium vitis. J Bacteriol 188:2173–2183
Kabbara S, Schmulling T, Papon N (2018) CHASEing cytokinin receptors in plants, bacteria, fungi, and beyond. Trends Plant Sci 23:179–181
Kim BS et al (2018) QStatin, a selective inhibitor of quorum sensing in Vibrio species. MBio 9:e02262–e02217
Kim Y et al (2010) Crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription. J Biol Chem 285:14020–14030
Lerat E, Moran NA (2004) The evolutionary history of quorum-sensing systems in bacteria. Mol Biol Evol 21:903–913
Lintz MJ, Oinuma K, Wysoczynski CL, Greenberg EP, Churchill ME (2011) Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc Natl Acad Sci USA 108:15763–15768
Mukherjee S, Bassler BL (2019) Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol 17:371–382
Papenfort K, Bassler BL (2016) Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588
Patankar AV, Gonzalez JE (2009) Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev 33:739–756
Patel HK, Suarez-Moreno ZR, Degrassi G, Subramoni S, Gonzalez JF, Venturi V (2013) Bacterial LuxR solos have evolved to respond to different molecules including signals from plants. Front Plant Sci 4:447
Rajput A, Kumar M (2017) In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria. Sci Rep 7:6969
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324
Schaefer AL et al (2016) A LuxR homolog in a cottonwood tree endophyte that activates gene expression in response to a plant signal or specific peptides. Mbio 7:e01101–e01116
Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG (2001) Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158–2163
Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB (2003) Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA 100:8951–8956
Subramoni S, Florez Salcedo DV, Suarez-Moreno ZR (2015) A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front Cell Infect Microbiol 5:16
Subramoni S et al (2011) Bacterial subfamily of LuxR regulators that respond to plant compounds. Appl Environ Microbiol 77:4579–4588
Subramoni S, Venturi V (2009) LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology 155:1377–1385
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Venturi V, Ahmer BM (2015) Editorial: LuxR solos are becoming major players in cell-cell communication in bacteria. Front Cell Infect Microbiol 5:89
Wang FF, Cheng ST, Wu Y, Ren BZ, Qian W (2017) A bacterial receptor PcrK senses the plant hormone cytokinin to promote adaptation to oxidative stress. Cell Rep 21:2940–2951
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Welsh MA, Blackwell HE (2016) Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol Rev 40:774–794
Whiteley M, Diggle SP, Greenberg EP (2017) Progress in and promise of bacterial quorum sensing research. Nature 551:313–320
Xu G, Yang S, Meng L, Wang BG (2018) The plant hormone abscisic acid regulates the growth and metabolism of endophytic fungus Aspergillus nidulans. Sci Rep 8:6504
Xu HY, Zhao YC, Qian GL, Liu FQ (2015) XocR, a LuxR solo required for virulence in Xanthomonas oryzae pv. oryzicola. Front Cell Infect Microbiol 5:37
Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ (2006) Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol 355:262–273
You L, Cox RS 3rd, Weiss R, Arnold FH (2004) Programmed population control by cell-cell communication and regulated killing. Nature 428:868–871
Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR (2010) A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol 400:847–864
Zhang L, Jia Y, Wang L, Fang R (2007) A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris. Mol Microbiol 65:121–136
Zhang RG et al (2002) Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971–974
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31570038).
Supporting information
Supplementary Table 1—Summary of characterized LuxR solo homologs in bacteria.
Supplementary Table 2—List of LuxR_Vha homologs in the TetR family.
Supplementary Fig. 1—Structure-based multiple sequence alignment (MSA) analysis of the LuxR_Vfi homologs.
Supplementary Fig. 2—Structure prediction and comparison of the LuxR_Vfi homologs.
Supplementary Fig. 3—The evolutionary conservation in LuxR_Vfi homologs estimated and visualized by the ConSurf server (Ashkenazy et al. 2016).
Supplementary Fig. 4—Structure-based MSA analysis of the LuxR_Vha homologs.
Supplementary Fig. 5—Structure prediction of the LuxR_Vha and comparison with the MDR regulators.
Supplementary Fig. 6—The evolutionary conservation in LuxR_Vha and MDR regulators estimated and visualized by the ConSurf server (Ashkenazy et al. 2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, G. Evolution of LuxR solos in bacterial communication: receptors and signals. Biotechnol Lett 42, 181–186 (2020). https://doi.org/10.1007/s10529-019-02763-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02763-6