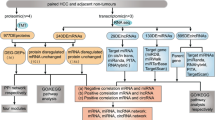
Abstract
Objective
To characterize the transcriptome profile of hepatocellular carcinoma (HCC) HepG2 cells treated with peptide 9R-P201 for further functional verification and HCC drug development.
Results
1557 mRNAs (1125 upregulated and 432 downregulated) and 881 lncRNAs (640 upregulated and 241 downregulated) with significant differential expression were identified using RNA-seq. The qRT-PCR results showed that the differential expression of several mRNAs and lncRNAs coincided with the RNA-seq results. Differentially expressed mRNAs and lncRNAs presented a significant difference in genomic characteristics but no preference under 9R-P201 treatment compared with control. The GO and KEGG functional enrichment analyses showed that differentially expressed mRNAs and lncRNAs remarkably enriched in cancer-related biological processes and signaling pathways. Finally, we screened out 33 TFs, 273 lncRNAs and 94 target genes with high degree interaction which were remarkably associated with the tumorigenesis and progression of cancers using betweenness centrality analysis.
Conclusion
These findings offer novel insights into the mechanism of 9R-P201 in HepG2 cells and provide new opportunities for the future 9R-P201-based drug development and the treatment of hepatocellular carcinoma.



Similar content being viewed by others
References
Alam T, Medvedeva YA, Jia H, Brown JB, Lipovich L, Bajic VB (2014) Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PLoS ONE 9:e109443
Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Beissbarth T, Speed TP (2004) GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20:1464–1465
Bi Z, Liu W, Ding R, Wu Y, Dou R, Zhang W, Yuan X, Liu X, Xiong L, Guo Z, Mao C (2017) A novel peptide, 9R-P201, strongly inhibits the viability, proliferation and migration of liver cancer HepG2 cells and induces apoptosis by down-regulation of FoxM1 expression. Eur J Pharmacol 796:175–189
Cui J, Huang J, Guo T, Wu F, Liang Z, Xiong L, Huang T, Zeng L, Liu N, Mao C (2014) Expression and selection of human foxm1c binding peptides and their inhibitions on MCF7 cancer cells. Int J Pept Res Ther 20:447–456
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789
Dong X, Chen K, Cuevas-Diaz Duran R, You Y, Sloan SA, Zhang Y, Zong S, Cao Q, Barres BA, Wu JQ (2015) Comprehensive identification of long non-coding RNAs in purified cell types from the brain reveals functional lncRNA in OPC fate determination. PLoS Genet 11:e1005669
Ghosh S, Sati S, Sengupta S, Scaria V (2015) Distinct patterns of epigenetic marks and transcription factor binding sites across promoters of sense-intronic long noncoding RNAs. J Genet 94:17–25
Han Y, Dong Q, Hao J, Fu L, Han X, Zheng X, Wang E (2016) RASSF4 is downregulated in nonsmall cell lung cancer and inhibits cancer cell proliferation and invasion. Tumour Biol 37:4865–4871
Hsieh TH, Hsu CY, Tsai CF, Chiu CC, Liang SS, Wang TN, Kuo PL, Long CY, Tsai EM (2016) A novel cell-penetrating peptide suppresses breast tumorigenesis by inhibiting beta-catenin/LEF-1 signaling. Sci Rep 6:19156
Huang JL, Zheng L, Hu YW, Wang Q (2014) Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis 35:507–514
Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ (2014) The UCSC Genome Browser database: 2014 update. Nucl Acid Res 42:D764–770
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Method 12:357–360
Kunz P, Sahr H, Lehner B, Fischer C, Seebach E, Fellenberg J (2016) Elevated ratio of MMP2/MMP9 activity is associated with poor response to chemotherapy in osteosarcoma. BMC Cancer 16:223
Liang H, Wang R, Jin Y, Li J, Zhang S (2016) MiR-422a acts as a tumor suppressor in glioblastoma by targeting PIK3CA. Am J Cancer Res 6:1695–1707
Liu K, Li X, Cao Y, Ge Y, Wang J, Shi B (2015) MiR-132 inhibits cell proliferation, invasion and migration of hepatocellular carcinoma by targeting PIK3R3. Int J Oncol 47:1585–1593
Melling N, Taskin B, Hube-Magg C, Kluth M, Minner S, Koop C, Grob T, Graefen M, Heinzer H, Tsourlakis MC, Izbicki J, Wittmer C, Huland H, Simon R, Wilczak W, Sauter G, Steurer S, Schlomm T, Krech T (2016) Cytoplasmic accumulation of ELAVL1 is an independent predictor of biochemical recurrence associated with genomic instability in prostate cancer. Prostate 76:259–272
Mora A, Donaldson IM (2011) iRefR: an R package to manipulate the iRefIndex consolidated protein interaction database. BMC Bioinf 12:455
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Rui Q, Xu Z, Yang P, He Z (2015) Long noncoding RNA expression patterns in lymph node metastasis in colorectal cancer by microarray. Biomed Pharmacother 75:12–18
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X, Yang Y, Xiao B, Guo J (2016) LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget 7:37812–37824
Shen H, Cheng H, Chen H, Zhang J (2017) Identification of key genes induced by platelet-rich plasma in human dermal papilla cells using bioinformatics methods. Mol Med Rep 15:81–88
Tang FH, Hsieh TH, Hsu CY, Lin HY, Long CY, Cheng KH, Tsai EM (2016) KRAS mutation coupled with p53 loss is sufficient to induce ovarian carcinosarcomas in mice. Int J Cancer 140:1860–1869
Wang G, Yang X, Li C, Cao X, Luo X, Hu J (2014) PIK3R3 induces epithelial-to-mesenchymal transition and promotes metastasis in colorectal cancer. Mol Cancer Ther 13:1837–1847
Zhang M, Chan MH, Tu WJ, He LR, Lee CM, He M (2013) Using the theory of coevolution to predict protein-protein interactions in non-small cell lung cancer. Chin J Cancer 32:91–98
Zhang X, Sha M, Yao Y, Da J, Jing D (2015) Increased B-type-natriuretic peptide promotes myocardial cell apoptosis via the B-type-natriuretic peptide/long non-coding RNA LSINCT5/caspase-1/interleukin 1beta signaling pathway. Mol Med Rep 12:6761–6767
Acknowledgements
This work was financially supported by the Program of the Fundamental Research Funds for the Central Universities of China (2682016YXZT04) and National Natural Science Foundation of China (No. 31200999).
Supporting information
Supplementary Table 1—The qRT-PCR primers of related genes and lncRNAs.
Supplementary Table 2—Differentially expressed mRNAs in Treatment vs. Control.
Supplementary Table 3—Differentially expressed lncRNAs in Treatment vs. Control.
Supplementary Table 4—The functinal enrichment results of differentially expressed lncRNAs.
Supplementary Table 5—The functinal enrichment results of differentially expressed mRNAs.
Supplementary Table 6—The transcription factor binding site analysis of differentially expressed lncRNAs.
Supplementary Table 7—The regulation and interaction relationship of differentially expressed lncRNAs.
Supplementary Table 8—The mutual interaction relationship of differentially expressed mRNAs.
Supplementary Fig. 1—Statistical analysis of clean reads from the control and treatment group.
Supplementary Fig. 2—The screening of the differentially expressed mRNAs and lncRNAs.
Supplementary Fig. 3—Comparative characteristics analysis of differentially expressed mRNAs and lncRNAs.
Supplementary Fig. 4—Comparative characteristics analysis of the all mRNAs and lncRNAs detected in transcriptome sequencing.
Supplementary Fig. 5—The GO function and KEGG pathway annotation analysis of differentially expressed mRNAs and lncRNAs target genes.
Supplementary Fig. 6—Differentially expressed mRNAs were significantly enriched in GO terms and KEGG signaling pathways closely related to cancer.
Supplementary Fig. 7—Cancer-related GO function and KEGG signaling pathway enrichment analysis of differentially expressed mRNAs and lncRNAs.
Supplementary Fig. 8—The regulatory network of TFs-lncRNAs-target genes.
Supplementary Fig. 9—The mutual interaction network of differentially expressed mRNAs.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, W., Ding, R., Zhang, Y. et al. Transcriptome profiling analysis of differentially expressed mRNAs and lncRNAs in HepG2 cells treated with peptide 9R-P201. Biotechnol Lett 39, 1639–1647 (2017). https://doi.org/10.1007/s10529-017-2407-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-017-2407-1