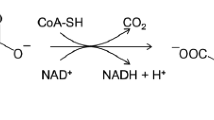
Abstract
Purpose of work
A pair of NAD+- and NADP+-dependent group III-alcohol dehydrogenases was characterized from the enterobacterium, Dickeya zeae, to expand our understanding of the distribution and biochemical properties of this interesting group of enzymes.
Two putative group III-alcohol dehydrogenases (ADHs) were identified in the genome of Dickeya zeae. Amino acid alignments and phylogenetic analysis revealed that Adh3.1 and Adh3.2 are only distantly related (~25 % identity at the protein level). Both proteins were purified to homogeneity after heterologous expression in E. coli. A specific activity of 1.8 U/mg was measured for the NAD+-dependent enzyme Adh3.1 with ethanol used as substrate, while NADPH-dependent Adh3.2 preferred butanal (29.1 U/mg) as substrate. Maximum activity for Adh3.1 was at 50 °C and pH 10 and for Adh3.2 at 70 °C and pH 6. Cell viability assays were used to confirm activity towards butanal and glyoxals. Biochemical characterization and phylogenetic analyses led to the hypothesis that Adh3.1 and Adh3.2 are probably the result of an ancient gene duplication event followed by functional diversification.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950
Hernandez-Tobias A, Julian-Sanchez A, Pina E, Riveros-Rosas H (2011) Natural alcohol exposure: is ethanol the main substrate for alcohol dehydrogenases in animals? Chem Biol Interact 191:14–25
Horn U, Strittmatter W, Krebber A, Knupfer U, Kujau M, Wenderoth R, Muller K, Matzku S, Pluckthun A, Riesenberg D (1996) High volumetric yields of functional dimeric miniantibodies in Escherichia coli, using an optimized expression vector and high-cell-density fermentation under non-limited growth conditions. Appl Microbiol Biotechnol 46:524–532
Jarboe LR (2011) YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 89:249–257
Lee C, Park C (2012) Development of a suicidal vector-cloning system based on butanal susceptibility due to an expression of YqhD aldehyde reductase. J Microbiol 50:249–255
Lee C, Kim I, Lee J, Lee KL, Min B, Park C (2010) Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J Bacteriol 192:4205–4214
Ma C, Zhang L, Dai J, Xiu Z (2010) Relaxing the coenzyme specificity of 1,3-propanediol oxidoreductase from Klebsiella pneumoniae by rational design. J Biotechnol 146:173–178
Marcal D, Rego AT, Carrondo MA, Enguita FJ (2009) 1,3-Propanediol dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity. J Bacteriol 191:1143–1151
Montella C, Bellsolell L, Perez-Luque R, Badia J, Baldoma L, Coll M, Aguilar J (2005) Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. J Bacteriol 187:4957–4966
Moon JH, Lee HJ, Park SY, Song JM, Park MY, Park HM, Sun J, Park JH, Kim BY, Kim JS (2011) Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor. J Mol Biol 407:413–424
Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC (2008) Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem 283:7346–7353
Quaglia D, Irwin JA, Paradisi F (2012) Horse liver alcohol dehydrogenase: new perspectives for an old enzyme. Mol Biotechnol 52:244–250
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Radianingtyas H, Wright PC (2003) Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol Rev 27:593–616
Sulzenbacher G, Alvarez K, Van Den Heuvel RH, Versluis C, Spinelli S, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M (2004) Crystal structure of E.coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J Mol Biol 342:489–502
Timpson LM, Liliensiek AK, Alsafadi D, Cassidy J, Sharkey MA, Liddell S, Allers T, Paradisi F (2012) A comparison of two novel alcohol dehydrogenase enzymes (ADH1 and ADH2) from the extreme halophile Haloferax volcanii. Appl Microbiol Biotechnol 97(1):195–203
Xiu ZL, Zeng AP (2008) Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl Microbiol Biotechnol 78:917–926
Acknowledgments
The authors thank Henning Piascheck for the help with the fermentation experiments. This work was funded by the Excellence Cluster in the Excellence Initative by the State of Hamburg “Fundamentals of Synthetic Biological Systems (SynBio)”.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Elleuche, S., Klippel, B., von der Heyde, A. et al. Comparative analysis of two members of the metal ion-containing group III-alcohol dehydrogenases from Dickeya zeae . Biotechnol Lett 35, 725–733 (2013). https://doi.org/10.1007/s10529-013-1137-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1137-2