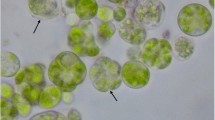
Abstract
A colony PCR technique was applied for both genomic and chloroplast DNA in the green microalgae Chlorella. Of five different lysis buffers, Chelex-100 was superior for DNA extraction, PCR and DNA storage. It also was insensitive to variations in cell density. The conditions established for an improved PCR formulation are applicable for screening of genetically-engineered transformants as well as bioprospecting of natural microalgal isolates. Besides multiple Chlorella species, we also demonstrate the efficacy of Chelex-100 for colony PCR with a number of other microalgal strains, including Chlamydomonas reinhardtii, Dunaliella salina, Nannochloropsis sp., Coccomyxa sp., and Thalassiosira pseudonana.




Similar content being viewed by others
References
Cao M, Fu Y, Guo Y, Pan J (2009) Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma 235:107–110
Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 25:294–306
Görs M, Schumann R, Hepperle D, Karsten U (2010) Quality analysis of commercial Chlorella products used as dietary supplement in human nutrition. J Appl Phycol 22:265–276
Hofmann MA, Brian DA (1991) Sequencing PCR DNA amplified directly from a bacterial colony. Biotechniques 11:30–31
Liang Y, Sarkany N, Cui Y (2009) Biomass and lipid productivities of Chlorella vulgaris under autotrophic, heterotrophic and mixotrophic growth conditions. Biotechnol Lett 31:1043–1049
Lv JM, Cheng LH, Xu XH, Zhang L, Chen HL (2010) Enhanced lipid production of Chlorella vulgaris by adjustment of cultivation conditions. Bioresour Technol 101:6797–6804
Rosenberg JN, Oyler GA, Wilkinson L, Betenbaugh MJ (2008) A green light for engineered algae: redirecting metabolism to fuel a biotechnology revolution. Curr Opin Biotechnol 19:430–436
van Zeijl CM, van de Kamp EH, Punt PJ, Selten GC, Hauer B, van Gorcom RF, van den Hondel CA (1998) An improved colony-PCR method for filamentous fungi for amplification of PCR-fragments of several kilobases. J Biotechnol 59:221–224
Verma D, Daniell H (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145:1129–1143
Walsh P, Metzger D, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Ward AC (1992) Rapid analysis of yeast transformants using colony-PCR. Biotechniques 13:350
Zimmermann J, Wiemann S, Voss H, Schwager C, Ansorge W (1994) Improved fluorescent cycle sequencing protocol allows reading nearly 1000 bases. Biotechniques 17:302, 304, 306
Acknowledgments
This work was supported by Chinese Natural Science Foundation For Distinguished Group (No. 50621063) and Graduate Innovation Project of Central South University (No. 2010bsxt05).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wan, M., Rosenberg, J.N., Faruq, J. et al. An improved colony PCR procedure for genetic screening of Chlorella and related microalgae. Biotechnol Lett 33, 1615–1619 (2011). https://doi.org/10.1007/s10529-011-0596-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-011-0596-6