Abstract
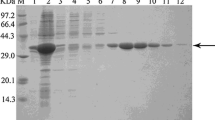
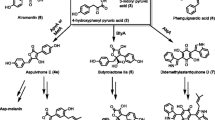
To elucidate the biotransformation from 5-oxomilbemycins A3 and A4 to milbemycins A3 and A4 in Streptomyces bingchengensis, the C5-ketoreductase gene (milF) was cloned using PCR with the specific primer designed from homologous nucleotide sequences. The C5-ketoreductase (MilF) was heterologously expressed in E. coli BL21 (DE3) as a His-tagged fusion protein. The characterization and biotransformation function of purified MilF was verified by in vitro enzyme assay. MilF is an NADPH-dependent reductase. The biotransformation products, analyzed by LC-APCI/MS, were identified as milbemycin A3 and milbemycin A4. MilF is thus present in Streptomyces bingchengensis and can transform 5-oxomilbemycins A3 and A4 to milbemycins A3 and A4. These findings are significant for understanding the biosynthetic pathway of milbemycins in Streptomyces bingchengensis and pave the way to obtain a producer strain of 5-oxomilbemycins directly by targeted milF disruption.



Similar content being viewed by others
References
De Bernardez Clark E, Schwarz EF, Rudolph R (1999) Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol 309:217–236
He YL, Sun YH, Liu TG, Zhou XF, Bai LQ, Deng ZX (2010) Cloning of the separate meilingmycin biosynthetic gene clusters through acyltransferase–ketoreductase (AT–KR) di-domain PCR amplification. Appl Environ Microbiol. doi:10.1128/AEM.02262–09
Ide J, Okazaki T, Ono M, Saito A, Nakagawa K, Naito S, Sato K, Tanaka K, Yoshikawa H, Ando M, Katsumi S, Matsumoto K, Toyama T, Shibano M, Abe M (1993) Milbemycin: discovery and development. Annu Rep Sankyo Res Lab 45:1–98
Ikeda H, Omura S (1997) Avermectin biosynthesis. Chem Rev 97:2591–2609
Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S (1999) Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Biochemistry 96:9509–9514
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics, 2nd edn. The John Innes Foundation, Norwich
Nonaka K, Kumasaka C, Okamoto Y, Maruyama F, Yoshikawa H (1999) Bioconversion of milbemycin-related compounds: biosynthetic pathway of milbemycins. J Antibiot 52:109–116
Nonaka K, Tsukiyama T, Okamoto Y et al (2000) New milbemycins from Streptomyces hygroscopicus subsp. aureolacrimosus: fermentation, isolation and structure elucidation. J Antibiot 53:694–704
Persson B, Krook M, Jornvall H (1991) Characteristics of short chain alcohol dehydrogenases and related enzymes. Eur J Biochem 200:537–543
Sambrook J et al (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Sun YH, Zhou XF, Tu GQ, Deng ZX (2003) Identification of a gene cluster encoding meilingmycin biosynthesis among multiple polyketide synthase contigs isolated from Streptomyces nanchangensis NS3226. Arch Microbiol 180:101–107
Wang XJ, Wang JD, Xiang WS (2009a) Three new milbemycin derivatives from Streptomyces bingchenggensis. J Asian Nat Prod Res 11:597–603
Wang XJ, Guo SL, Guo WQ et al (2009b) Role of nsdA in negative regulation of antibiotic production and morphological differentiation in Streptomyces bingchengensis. J Antibiot 62:309–313
Xiang WS, Wang JD, Wang XJ, Zhang J (2007) Two new β-class milbemycins from Streptomyces bingchenggensis: fermentation, isolation, structure elucidation and biological properties. J Antibiot 60(6):351–356
Xiang WS, Wang JD, Wang XJ et al (2008) New seco-milbemycins from Streptomyces bingchenggensis: fermentation, isolation and structure elucidation. J Antibiot 61:27–32
Acknowledgements
This study was supported by the National Key Technology R&D Program (No. 2006BAD31B02), the Program for New Century Excellent Talents in University (No. NCET-08-0668, 1154-NCET-002), the Outstanding Youth Foundation of Heilongjiang Province (No. JC200706), the National Natural Science Foundation of China (No. 30971937), the National Key Project for Basic Research (No. 2010CB126102), the Program for New Teachers in University (No. 20092325120007) and the Innovative Program for Postgraduates in Heilongjiang Province (No. YJSCX2009-149HLJ).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, XJ., Wang, CQ., Sun, XL. et al. 5-ketoreductase from Streptomyces bingchengensis: overexpression and preliminary characterization. Biotechnol Lett 32, 1497–1502 (2010). https://doi.org/10.1007/s10529-010-0320-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-010-0320-y