Abstract
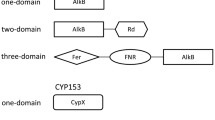
Rhodococcus sp. TMP2 is an alkane-degrading strain that can grow with a branched alkane as a sole carbon source. TMP2 degrades considerable amounts of pristane at 20°C but not at 30°C. In order to gain insights into microbial alkane degradation, we characterized one of the key enzymes for alkane degradation. TMP2 contains at least five genes for membrane-bound, non-heme iron, alkane hydroxylase, known as AlkB (alkB1–5). Phylogenetical analysis using bacterial alkB genes indicates that TMP2 is a close relative of the alkane-degrading bacteria, such as Rhodococcus erythropolis NRRL B-16531 and Q15. RT-PCR analysis showed that expressions of the genes for AlkB1 and AlkB2 were apparently induced by the addition of pristane at a low temperature. The results suggest that TMP2 recruits certain alkane hydroxylase systems to utilize a branched alkane under low temperature conditions.




Similar content being viewed by others
References
Baek KH, Yoon BD, Lee IS, Oh HM, Kim HS (2006) Biodegradation of aliphatic aromatic hydrocarbons by Nocardia sp. H17-1. Geomicrobiol J 23:253–259
Kunihiro N, Haruki M, Takano K, Morikawa M, Kanaya S (2005) Isolation and characterization of Rhodococcus sp. strains TMP2 and T12 that degrade 2,6,10,14-tetramethylpentadecane (pristane) at moderately low temperatures. J Biotechnol 115:129–136
Lang S, Philp JC (1998) Surface-active lipids in rhodococci. Antonie Van Leeuwenhoek 74:59–70
Nakajima K, Sato A, Takahara Y, Iida T (1985) Microbial oxidation of isoprenoid alkanes, phytane, norpristane and farnesane. Agric Biol Chem 49:1993–2002
Rapp P, Gabriel-Jürgens LH (2003) Degradation of alkanes and highly chlorinated benzenes, and production of biosurfactants, by a psychrophilic Rhodococcus sp. and genetic characterization of its chlorobenzene dioxygenase. Microbiology 149:2879–2890
Rehm H, Reiff I (1981) Mechanism and occurrence of microbial oxidation of long chain alkanes. Adv Biochem Eng 19:175–215
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sanin SL, Sanin FD, Bryers JD (2003) Effect of starvation on adhesive properties of xenobiotic degrading bacteria. Process Biochem 38:909–914
Sharma SL, Pant A (2000) Biodegradation and conversion of alkanes and crude oil by a marine Rhodococcus sp. Biodegradation 11:289–294
Staijien IE, van Beilen JB, Witholt B (2000) Expression, stability and performance of the three-component alkane mono-oxygenase of Pseudomonas oleovorans in Escherichia coli. Eur J Biochem 267:1957–1965
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
van Beilen JB, Funhoff EG (2005) Expanding the alkane oxygenase toolbox: new enzymes and applications. Curr Opin Biotechnol 16:308–314
van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21
van Beilen LB, Smits TH, Whyte LG, Schorcht S, Rothlisberger M, Plaggemeier T, Engesser KH, Witholt B (2002) Alkane hydroxylase homologues in Gram-positive strains. Environ Microbiol 4:676–682
Whyte LG, Slagman SJ, Pietrantonio F, Bourbonniere L, Koval SF, Lawrence JR, Inniss WE, Greer CW (1999) Physiological adaptations involved in alkane assimilation at a low temperature by Rhodococcus sp. strain Q15. Appl Environ Microbiol 65:2961–2968
Whyte LG, Smits TH, Labbe D, Witholt B, Greer CW, van Beilen JB (2002) Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl Environ Microbiol 68:5933–5942
Acknowledgements
This work was supported by New Energy and Industrial Technology Development Organization (NEDO), Institute for Fermentation, Osaka (IFO), and a Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (JSPS) to MM.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Takei, D., Washio, K. & Morikawa, M. Identification of alkane hydroxylase genes in Rhodococcus sp. strain TMP2 that degrades a branched alkane. Biotechnol Lett 30, 1447–1452 (2008). https://doi.org/10.1007/s10529-008-9710-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9710-9