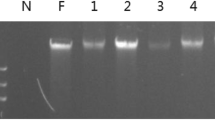
Feces are increasingly used as sources of DNA for genetic and ecological research. This paper describes a new method for isolation of DNA from animal feces. This method combines multiple purification steps, including pretreatment with ethanol and TE, an inhibitor-absorber made of starch, the CTAB method, the phenol–chloroform extraction method, and the guanidinium thiocyanate-silica method. The new method is efficient according to PCR results of 585 fecal samples from 23 species and costs much less than the commercial kits. The protocol can be tailored to the specific purpose of examining different diets of animals and can be performed with routine laboratory reagents.



Similar content being viewed by others
REFERENCES
Bianchini, F., Caderni, G., Dolara, P., Fantetti, L., and Kriebel, D. (1989). Effect of dietary fat, starch and cellulose on fecal bile acids in mice. J. Nutr. 119:1617–1624.
Cheah, P. Y., and Bernstein, H. (1990). Colon cancer and dietary fiber: Cellulose inhibits the DNA-damaging ability of bile acids. Nutr. Cancer 13:51–57.
Constable, J. J., Packer, C., Colins, D. A., and Pusy, A. E. (1995). Nuclear DNA from primate dung. Nature 373:393.
Deuter, R., Pietsch, S., Hertel, S., and Müller, O. (1995). A method for preparation of fecal DNA suitable for PCR. Nucleic Acids Res. 23:3800–3801.
Edwards, U., Rogall, T., Blocker, H., Emde, M., and Boettger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes: Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843–7853.
Eggert, L. S., Eggert J. A., and Woodruff, D. S. (2003). Estimating population sizes for elusive animals: The forest elephants of Kakum National Park, Ghana. Mol. Ecol. 12:1389–1402.
Ernest, H. B., Penedo, M. C., May, B. P., Syvanen, M., and Boyce, W. M. (2000). Molecular tracking of mountain lions in the Yosemite valley region in California: Genetic analysis using microsatellites and fecal DNA. Mol. Ecol. 9:433–441.
Fang, G., Hammar, S., and Grumet, R. A. (1992). Quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques 13:52–54.
Fernando, P., Vidya, T. N. C., and Melnick, D. J. (2001). Isolation and characterization of tri- and tetranucleotide microsatellite loci in the Asian elephant, Elephas Maximus. Mol. Ecol. Notes 1:232–233.
Frantz, A. C., Pope, L. C., Carpenter, P. J., Roper, T. J., Wilson, G. J., Delahay, R. J., and Burke, T. (2003). Reliable microsatellite genotyping of the Eurasian badger Meles meles using fecal DNA. Mol. Ecol. 12:1649–1661.
Höss, M., and Pääbo, S. (1993). DNA extraction from Pleistocene bones by a silica-based purification method. Nucleic Acids Res. 21:3913–3914.
Höss, M., Kohn, M., Pääbo, S., Knauer, F., and Schroder, W. (1992). Excrement analysis by PCR. Nature 359:199.
Irwin, D. M., Kocher, T. D., and Wilson, A. C. (1991). Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32:128–144.
Jones, A. S., and Walker, R. T. (1963). Isolation and analysis of the deoxyribonucleic acid of Mycoplasma mycoides var. Capri. Nature 198:588–589.
Kocher, T. D., Thomas, W. K., Meyer, A., Edwards, S. V., Pääbo, S., and Villablanca, F. X. (1989). Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. U.S.A. 86:6196–6200.
Kohn, M. H., and Wayne, R. K. (1997). Facts from feces revisited. TREE 12:223–327.
Lantz, P. G., Tjerneld, F., Hahn-Hagerdal, B., and Radstrom, P. (1996). Use of aqueous two-phase systems in sample preparation for polymerase chain reaction-based detection of microorganisms. J. Chromatogr. B. Biomed. Appl. 680:165–170.
Li, M., Gong, J., Cottrill, M., Yu, H., de Lange, C., Burton, J., and Topp, E. (2003). Evaluation of QIAampR DNA Stool Mini Kit for ecological studies of gut microbiota. J. Microbiol. Meth. 54:13–17.
Lu, Z., Johnson, W. E., Menotti-Raymond, M., Yuhki, N., Martenson, J. S., Mainka, S., Huang, S. Q., Zheng, Z., Li, G., Pan, W., Mao, X., and O’Brien, S. J. (2001). Patterns of genetic diversity in remaining giant panda populations. Conserv. Biol. 15:1596–1607.
Machiels, B. M., Ruers, T., Lindhout, M., Hlavaty, T., Bang, D. D., Somers, V. A. M. C., Baeten, C., von Meyenfeldt, M., and Thunnissen, F. B. J. M. (2000). New protocol for DNA extraction of stool. Biotechniques 28:286–290.
Monteiro, L., Bonnemaison, D., Vekris, A., Petry, K. G., Bonnet, J., Vidal, R., Cabrita, J., and Mégraud, F. (1997). Complex polysaccharides as PCR inhibitors in feces: Helicobacter pyori model. J. Clin. Microbiol. 35:995–998.
Murphy, M. A., Waits, L. P., and Kendall, K. C. (2000). Quantitative evaluation of fecal drying methods for brown bear DNA analysis. Wildlife Soc. B 28:951–957.
Reed, J. Z., Tollit, D. J., Thompson, P. M., and Amos, W. (1997). Molecular scatology: The use of molecular genetic analysis to assign species, sex and individual identity to seal feces. Mol. Ecol. 6:225–234.
Taberlet, P., Griffin, S., Goossens, B., Questiau S., Manceau, V., Escaravage, N., Waits, L. P., and Bouvet, J. (1996). Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 24:3189–3194.
Taberlet, P., and Luikart, G. (1999). Non-invasive genetic sampling and individual identification. Biol. J. Linn. Soc. 68:41–55.
Walsh, P. S., Metzger, D. A., and Higuchi, R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513.
ACKNOWLEDGMENTS
This research was funded by the CAS Innovative Research International Partnership Project (CXTDS2005-4) and State Forestry Administration of China. Special thanks are given to Mr. Liu Zhijin, Ms. Zeng Yan, Ms. Zhang Fangfang, Mr. Zhu Lifeng, and Ms. Liu Qing for their lab work. We are also grateful to Dr. Zhang Zejun, who helped with sampling.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, BW., Li, M., Ma, LC. et al. A Widely Applicable Protocol for DNA Isolation from Fecal Samples. Biochem Genet 44, 494–503 (2006). https://doi.org/10.1007/s10528-006-9050-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-006-9050-1