Abstract
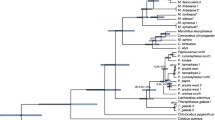
To study the phylogenetic relationships of the macaques, five gene fragments were sequenced from 40 individuals of eight species: Macaca mulatta, M. cyclopis, M. fascicularis, M. arctoides, M. assamensis, M. thibetana, M. silenus, and M. leonina. In addition, sequences of M. sylvanus were obtained from Genbank. A baboon was used as the outgroup. The phylogenetic trees were constructed using maximum-parsimony and Bayesian methods. Because five gene fragments were from the mitochondrial genome and were inherited as a single entity without recombination, we combined the five genes into a single analysis. The parsimony bootstrap proportions we obtained were higher than those from earlier studies based on the combined mtDNA dataset. Excluding M. arctoides, our results are generally consistent with the classification of Delson (1980). Our phylogenetic analyses agree with earlier studies suggesting that the mitochondrial lineages of M. arctoides share a close evolutionary relationship with the mitochondrial lineages of the fascicularis group of macaques (and M. fascicularis, specifically). M. mulatta (with respect to M. cyclopis), M. assamensis assamensis (with respect to M. thibetana), and M. leonina (with respect to M. silenus) are paraphyletic based on our analysis of mitochondrial genes.
Similar content being viewed by others
References
Abegg, C., and Thierry, B. (2002). Macaque evolution and dispersal in insular south-east Asia. Biol. J. Linn. Soc. 75:555–576.
Collura, R. V., and Stewart, C. B. (1995). Insertions and duplications of mitochondrial DNA in the nuclear genomes of Old World monkeys and hominoids. Nature 378:486–489.
Cronin, J. E., Cann, R., and Sarich, V. M. (1980). Molecular evolution and systematics of the genus Macaca. In Lindburg, D. G. (ed.), The macaques: Studies in ecology behavior and evolution. Van Nostrand Reinhold Co., New York, pp. 31–51.
Deinard, A., and Smith, D. (2001). Phylogenetic relationships among the macaques: Evidence from the nuclear locus NRAMP1. J. hum. Evol. 41:45–59.
Delson, E. (1980). Fossil macaques phyletic relationships and a scenario of development. In Lindburg, D. G. (ed.), The macaques: Studies in ecology behavior and evolution, Van Nostrand Reinhold Co., New York, pp. 10–30.
Dewoody, J. A., Chesser, R. K., and Barker, R. J. (1999). A translocated mitochondrial cytochrome b pseudogene in voles (Rodentia: Microtus). J. Mol. Evol. 48:380–382.
Fa, J. E. (1989). The genus Macaca: A review of taxonomy and evolution. Mammal. Rev. 19:45–81.
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791.
Fooden, J. (1976). Provisional classification and key to living species of macaques (Primates: Macaca). Folia Primatol 25:225–236.
Fooden, J. (1982). Taxonomy and evolution of the sinica group of macaques: 3. species and subspecies accounts of Macaca assamensis. Fieldiana Zool 10:1–52.
Fooden, J. (1988). Taxonomy and evolution of the sinica group of macaques: 6. Interspecific comparisons and synthesis. Fieldiana Zool New Series 45:1–44.
Fooden, J., and Lanyon, S. M. (1989). Blood protein allele frequencies and phylogenetic relationships in Macaca: A review. Am. J. Primatol 17:209–241.
Groves, C. P. (2001). Primate Taxonomy. Smithsonian Institution Press, Washington DC.
Hayasaka, K., Fujii, K., and Horai, S. (1996). Molecular phylogeny of macaques: Implications of nucleotide sequences from an 896-base pair region of mitochondrial DNA. Mol. Biol. Evol. 13:1044–1053.
Hill, W. C. O. (1970). Primates: Comparative anatomy and taxonomy 8. Cynopithecinae: Papio, Mandrillus, Theropithecus. Edinburgh: Edinburgh University Press.
Hill, W. C. O. (1974). Primates: Comparative anatomy and taxonomy 7. Cynopithecinae: Cercocebus, Macaca, Cynopithecus. Edinburgh: Edinburgh University Press.
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182–192.
Hoelzer, G. A., Hoelzer, M. A., and Melnick, D. J. (1992). The evolution history of the sinica group of macaque monkeys as revealed by mtDNA restriction site analysis. Mol. Phylogenet Evol. 1:215–222.
Huelsenbeck, J. P., and Hillis, D. M. (1993). Success of phylogenetic methods in the four-taxon case. Syst. Biol. 42:247–264.
Huelsenbeck, J. P., and Ronquist, F. R. (2001). MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755.
Huelsenbeck, J. P., Bull, J. J., and Cunningham, W. (1996). Combining data in phylogenetic analysis. Trends Ecol. Evol. 11:152–158.
Kumar, S., Tamura, K., Jakobsen, I. B., and Nei, M. (2001). MEGA (molecular evolutionary genetics analysis) software project. (http://www.megasoftware.net/), visited 26 September 2003.
Li, Q., and Zhang, Y. (2004). A Molecular Phylogeny of Macaca Based on Mitochondrial Control Region Sequences. Zool. Res. 25:385–390.
Lü, X., Fu, Y., and Zhang, Y. (2002). Evolution of mitochondrial cytochrome b pseudogene in genus Nycticebus. Mol. Biol. Evol. 19:2337–2341.
Melnick, D. J., Hoelzer, G. A., Absher, R., and Ashley, M. V. (1993). MtDNA diversity in rhesus monkeys reveals overestimates of divergence time and paraphyly with neighboring species. Mol. Biol. Evol. 10:282–295.
Melnick, D. J., and Kidd, K. K. (1985). Genetic and evolutionary relationships among Asian macaques. Int. J. Primatol. 6:123–160.
Morales, J., and Melnick, D. J. (1998). Phylogenetic relationships of the macaques (Cercopithecidae: Macaca) as revealed by high resolution restriction site mapping of mitochondrial ribosomal genes. J. Hum. Evol. 34:1–23.
Moreiro, M. A. M., and Seuanez, H. N. (1999). Mitochondrial pseudogenes and phyletic relationships of Cebuella and Callithrix (Platyrrhini Primates). Primates 40:353–364.
Murphy, W. J., Eizirik, E., O'Brien, S. J., Madsen, O., Scally, M., Douady, C. J., Teeling, E., Ryder, O. A., Stanhope, M. J., de Jong, W. W., and Springer, M. S. (2001). Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348–2351.
Napier, J. R., and Napier, P. M. (1967). A Handbook of Living Primates, Academic press, London/New York.
Naylor, G. J., Collins, T. M., and Brown, W. M. (1995). Hydrophobicity and phylogeny. Nature 373:565–566.
Nei, M. (1991). Relative efficiencies of different tree-making methods for molecular data. In Miyamoto, M. M., and Cracraft, J. (eds.), Phylogenetic Analysis of DNA Sequences. Oxford University, New York, pp. 90–128.
Nozawa, K., Shotake, T., Ohkura, Y., and Tanabe, Y., (1977). Genetic variations within and between species of Asian macaques. Jpn. J. Genet 52:15–30.
Pocock, R. I. (1926). The external characters of the Catarrhine monkeys and apes. Proc. Zool. Soc. London 1479–1579.
Posada, D., and Crandall, K. A. (1998). Modeltest: Testing the model of DNA substitution. Bioinformatics 14:817–818.
Springer, S., and Douzery, E. (1996). Secondary structure and patterns of evolution among mammalian mitochondrial 12S rRNA molecules. J. Mol. Evol. 43:357–373.
Swofford, D. L. (2001). PAUP* Phylogenetic Analysis Using Parsimony (* and other methods) 4.0b8a. Sinauer Sunderland MA.
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526.
Tanaka, T., and Takenaka, O. (1996). Phylogenetic relationship of the genus Macaca inferred from DNA sequence. In International Symposium: Evolution of Asian Primates, Freude and Kyoto University Primate Research Institute, Inuyama, Aichi, Japan, p. 10.
Tosi, A. J., Morales, J. C., and Melnick, D. J. (2000). Comparison of Y chromosome and mtDNA phylogenies leads to unique inferences of macaque evolutionary history. Mol. Phylogenet. Evol. 17:133–144.
Tosi, A. J., Morales, J. C., and Melnick, D. J. (2002). Y-chromosome and mitochondrial markers in Macaca fascicularis indicate introgression with Indochinese M. mulatta and a biogeographic barrier in the Isthmus of Kra. Int. J. Primatol. 23(1):161–178.
Tosi, A. J., Disotll, T. R., Morales, J. C., and Melnick, D. J. (2003a). Cercopithecine Y-chromosome data provide a test of competing morphological evolutionary hypotheses. Mol. Phylogenet. Evol. 27:510–521.
Tosi, A. J., Morales, J. C., and Melnick, D. J. (2003b). Paternal maternal and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57(6):1419–1435.
Wang, Y. (2003). A Complete Checklist of Mammal Species and Subspecies in China, a Taxonomic and Geographic Reference. China forestry publishing house, Beijing.
Wolstenholme, D. R. (1992). Animal mitochondrial DNA: Structure and evolution. In Wolstenholme, D. R. and Jeon, K. W. (eds.) “Mitochondrial genomes”, International Review of Cytology, Academic Press, San Diego, 141:173–216.
Yang, Z. (1994). Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 39:306–314.
Zhang, D. X., and Hewitt, G. M. (1996). Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol. Evol. 11:247–251.
Zhang, Y., Quan, G., Zhao, T., and Southwick, C. (1991). Distribution of Macaques (Macaca) in China. Acta. Theriologica Sinica. 11:171–185.
Zhang, Y., and Shi, L. (1993). Phylogenetic relationships of macaques as inferred from restriction endonuclease analysis of mitochondrial DNA. Folia Primatol. 60:7–17.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, QQ., Zhang, YP. Phylogenetic Relationships of the Macaques (Cercopithecidae: Macaca), Inferred from Mitochondrial DNA Sequences. Biochem Genet 43, 375–386 (2005). https://doi.org/10.1007/s10528-005-6777-z
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10528-005-6777-z