Abstract
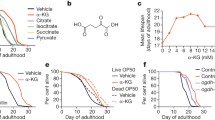
Dietary restriction is one of the several ways which could putatively extend organisms’ lifespan, ranging from Saccharomyces cerevisiae to rodents, by activating the AMP-activated protein kinase (AMPK), an ATP/AMP sensor. Extensive researches have shown that aging reduces sensibility of AMPK and eventually causes energy imbalance in cells. Research in mammals’ AMPK depicts that this signaling molecule could control autophagy, improve cellular stress resistance and suppress inflammatory responses. Hence, in this study we performed a drug repurposing of 1908 FDA-approved drugs in order to discover putative safe activators of AMPK and to find new applications for existing drugs. For this purpose, FDA-approved drugs were screened by virtual screening and the ligand–protein interactions were carefully inspected. Moreover, through MM/PBSA analysis, the binding affinity of hit compounds in γ and αβ binding sites were investigated. As Cangrelor, Nacitentan, Levoleucovorin and Glisoxepide had lower binding affinities; we predicted that they would probably prove to be more potential activators than C2. However, hit-compounds in αβ binding site, exhibited higher unfavorable binding affinity. Hence, present findings can prove to be valuable for discovering new activators for AMPK.







Similar content being viewed by others
References
Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1
Calvert S, Tacutu R, Sharifi S, Teixeira R, Ghosh P, Magalhães JP (2016) A network pharmacology approach reveals new candidate caloric restriction mimetics in C. elegans. Aging Cell 15:256–266
Cameron KO, Kurumbail RG (2016) Recent progress in the identification of adenosine monophosphate-activated protein kinase (AMPK) activators. Bioorganic Med Chem Lett 26:5139–5148
Cantó C et al (2009) AMPK regulates energy expenditure by modulating NAD + metabolism and SIRT1 activity. Nature 458:1056
Cavalla D (2013) Predictive methods in drug repurposing: gold mine or just a bigger haystack? Drug Discov Today 18:523–532
Chen L et al (2013) Conserved regulatory elements in AMPK. Nature 498:E8
Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA (1998) Functional domains of the α1 catalytic subunit of the AMP-activated protein kinase. J Biol Chem 273:35347–35354
Dalle Pezze P et al (2016) A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun 7:13254
Ganesan A, Coote ML, Barakat K (2017) Molecular dynamics-driven drug discovery: leaping forward with confidence. Drug Discov Today 22:249–269
Gupta SC, Sung B, Prasad S, Webb LJ, Aggarwal BB (2013) Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol Sci 34:508–517
Hall JA, Dominy JE, Lee Y, Puigserver P (2013) The sirtuin family’s role in aging and age-associated pathologies. J Clin Investig 123:973–979. https://doi.org/10.1172/JCI64094
Hardie DG, Hawley SA (2001) AMP-activated protein kinase: the energy charge hypothesis revisited. BioEssays 23:1112–1119
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem 32:866–877
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kumari R, Kumar R, Consortium OSDD, Lynn A (2014) g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Info Model 54:1951–1962
Li X et al (2015) Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res 25:50
Ma D-L, Chan DS-H, Leung C-H (2013) Drug repositioning by structure-based virtual screening. Chem Soc Rev 42:2130–2141
Maswood N et al (2004) Caloric restriction increases neurotrophic factor levels and attenuates neurochemical and behavioral deficits in a primate model of Parkinson’s disease. Proc Natl Acad Sci 101:18171–18176
Meng X-Y, Zhang H-X, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput-Aided Drug Des 7:146–157
Pronk S et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Rana S, Blowers EC, Natarajan A (2014) Small molecule adenosine 5′-monophosphate activated protein kinase (AMPK) modulators and human diseases. J Med Chem 58:2–29
Salminen A, Kaarniranta K (2012) AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res Rev 11:230–241
Sanders MJ, Ali ZS, Hegarty BD, Heath R, Snowden MA, Carling D (2007) Defining the mechanism of activation of AMP-activated protein kinase by the small molecule A-769662, a member of the thienopyridone family. J Biol Chem 282:32539–32548
Steinberg GR, Kemp BE (2009) AMPK in health and disease. Physiol Rev 89:1025–1078
Townley R, Shapiro L (2007) Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science 315:1726–1729
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Tsui V, Case DA (2000) Theory and applications of the generalized born solvation model in macromolecular simulations. Biopolymers 56:275–291
Wang Z et al (2016) Comprehensive evaluation of ten docking programs on a diverse set of protein–ligand complexes: the prediction accuracy of sampling power and scoring power. Phys Chem Chem Phys 18:12964–12975
Xiao B et al (2013) Structural basis of AMPK regulation by small molecule activators. Nat Commun 4:3017
Xin F-J, Wang J, Zhao R-Q, Wang Z-X, Wu J-W (2013) Coordinated regulation of AMPK activity by multiple elements in the α-subunit. Cell Res 23:1237
Acknowledgement
The authors are grateful to Ms. Shady Sheybani for her help to prepare some of the figures. This investigation was supported by a grant number 96-1206 from Golestan University, Gorgan, Iran.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10522_2018_9744_MOESM1_ESM.jpg
Model of AMPK regulation by dietry restriction and its effect on downstream proteins. Factors having negetive effects on longevity are coloured red and positive factors are shown in green. Supplementary material 1 (JPEG 159 kb)
10522_2018_9744_MOESM2_ESM.png
Heterotrimeric AMPK structure and the ligand binding site: blue, yellow and purple colors represent α, β and γ subunits, respectively. Supplementary material 2 (PNG 6836 kb)
10522_2018_9744_MOESM3_ESM.png
Backbone RMSD values of protein–ligand complexes of the γ site (A) and αβ site (B) during 10 ns of MD simulation. Supplementary material 3 (PNG 1224 kb)
10522_2018_9744_MOESM4_ESM.png
RMSF values of the residue in the presence of hit compound during 10 ns of MD simulation at γ binding site. Supplementary material 4 (PNG 1023 kb)
10522_2018_9744_MOESM5_ESM.png
RMSF values of the residue in the presence of hit compound during 10 ns of MD simulation at αβ binding site. Supplementary material 5 (PNG 724 kb)
10522_2018_9744_MOESM6_ESM.png
Hotspot residues and hydrogen bonds of hit compounds at γ active site of AMPK. (A) C2, (B) AMP, (C) Cangrelor, (D) Nacitentan, (E) Levoleucovorin and (F) Glisoxepide. Supplementary material 6 (PNG 2693 kb)
10522_2018_9744_MOESM7_ESM.png
Hotspot residues and hydrogen bonds of hit compounds at αβ active site of AMPK. (A) A769662, (B) Ceftolozane, (C) Ceftriaxone and (D) Levomefolic acid. Supplementary material 7 (PNG 1495 kb)
Rights and permissions
About this article
Cite this article
Mofidifar, S., Sohraby, F., Bagheri, M. et al. Repurposing existing drugs for new AMPK activators as a strategy to extend lifespan: a computer-aided drug discovery study. Biogerontology 19, 133–143 (2018). https://doi.org/10.1007/s10522-018-9744-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10522-018-9744-x