Abstract
In mammals, NO•, a signaling molecule is implicated in the regulation of vasodilation, neurotransmission and immune response. It is believed that NO• is a signaling molecule also in unicellular organism like yeast and may be involved in the regulation of apoptosis and sporulation. It has been reported that NO• is produced during chronological aging (CA) leading to an increase of the superoxide level, which in turn mediates apoptosis. Since this conclusion was based on indirect measurements of NO• by the Griess reaction, the role of NO• signaling during CA in the yeast remains uncertain. We investigated this issue more precisely using different genetic and biochemical methodologies. We used cells lacking the factors influencing nitrosative stress response like flavohemoglobin metabolizing NO•, S-nitrosoglutathione reductase metabolizing S-nitrosoglutathione and the transcription factor Fzf1p mediating NO• response. We measured the standard parameters describing CA and found an elevation in the superoxide level, percentage of death cells, the level of TUNEL positive cells and a decrease in proliferating potential. These observations showed no significant differences between wild type cells and the disruptants except for a small elevation of the superoxide level in the Δsfa1 mutant. The intracellular NO• level and flavohemoglobin expression decreased rather than increased during CA. Products of general nitrogen metabolism and protein tyrosine nitration were slightly decreased during CA, the magnitude of changes showing no differences between the wild type and the mutant yeast. Altogether, our data indicate that apoptosis during yeast CA is mediated by superoxide signaling rather than NO• signaling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The yeast Saccharomyces cerevisiae has become a very important model organism to study biochemistry and molecular biology of mammalian cells and to answer fundamental questions concerning the mechanisms of many processes like aging (Kaeberlein 2010). Thanks to the ease of genetic manipulation, its small, well-known genome and short life cycle, the yeast is widely used as an experimental model (Gershon and Gershon 2000). Additionally, owing to yeast asymmetric cell division, it is possible to determine yeast replicative lifespan (RLS), i. e., the number of daughter cells produced by a mother cell before senescence (Mortimer and Johnston 1959). Replicative aging of S. cerevisiae is believed to be a model of aging of mitotically active cells of higher eukaryotes. RLS is measured by counting daughter cells, which can be easily distinguished from mother cells under a microscope (Kaeberlein 2010). Moreover, chronological aging of the yeast (CA) is considered as a model of aging of post-mitotic cells of higher eukaryotes (Chen et al. 2005; MacLean et al. 2001). Chronological lifespan (CLS) is defined as the lifespan of cells in a non-dividing state in a stationary culture (Longo et al. 1996; Fabrizio and Longo 2003).
It has been shown that several genetic and environmental factors modulating CLS in yeast, like calorie restriction, Sch9 (S6 kinase) and TOR pathway, can also influence aging in other organisms (worms, flies, and mice) (Kennedy et al. 2007; Yan et al. 2007). A number of longevity-associated genes (LAGs) was revealed by genetic manipulation in yeast, worms, flies, and mice; they are highly evolutionary conserved across all these model organisms (Budovsky et al. 2007; de Magalhaes et al. 2009). Additionally, the model of yeast CA system has allowed to detect anti-aging compounds (like spermidine or rapamycin) which are also efficient in higher eukaryotes (Eisenberg et al. 2009; Powers et al. 2006; Harrison et al. 2009; Bjedov et al. 2010). Despite many studies identifying multiple molecular causes of aging like oxidative stress, senescence factors, telomere shortening, genomic instability, epigenetic changes or proteotoxicity (Harman 1956; Harley et al. 1992; Campisi 2005; Dolle and Vijg 2002; Cohen et al. 2006), the primary cause of aging of the human has not yet been determined.
The most commonly used protocol for studying CA in the yeast has been proposed by Longo (Fabrizio and Longo 2003). Cells cultured in liquid media up to carbon source depletion enter a non-dividing state. Then they are transferred to fresh medium which allows to monitor their ability to continue mitotic growth (expressed mainly as a colony forming units or CFUs) (Fabrizio and Longo 2003; Longo et al. 1996). Moreover, yeast CA can be study both during a hypo-metabolic phase (stationary phase) when cells are kept in water and during a high-metabolic phase (post-diauxic phase) when cells are cultured in synthetic dextrose complete (SDC) medium (Fabrizio and Longo 2003). Using SDC medium, a link between stress resistance and longevity was found and evidence for conservation of pathways modulating lifespan of evolutionally distinct eukaryotes was provided (Fabrizio et al. 2001).
It is suggested that CA in the yeast is mainly mediated by oxidative stress since markers of oxidative stress are increased during CA and cells lacking antioxidant machinery demonstrate reduced CLS (Longo et al. 1996; Fabrizio and Longo 2003; Jakubowski et al. 2000; Herker et al. 2004). Additionally, reactive oxygen species (ROS) may take part in the regulation of apoptosis during CA in the yeast which allows for the survival of the fittest and best adapted cells at the expense of ROS-damaged and old cells (Herker et al. 2004; Fabrizio et al. 2004).
ROS are regulated and controlled by antioxidant defenses and scavenging enzymes (Thorpe et al. 2004). Nevertheless, ROS are thought to be something more than unwanted by-products of metabolism (de Magalhaes and Church 2006; Bartosz 2009). ROS are considered as signaling molecules regulating many biological processes (Esposito et al. 2004). In mammals, a plethora of extracellular stimuli like growth factors stimulate a transient increase in intracellular ROS levels (Berner and Stern 2004). ROS may modulate redox reactions and active sites of enzymes thus they can affect the activity of transcription factors like NF-κB, JUN, and FOS (Remacle et al. 1995). By modulating protein tyrosine phosphatase activity, ROS may influence cellular growth and proliferation (Chiarugi and Cirri 2003). In mammals, ROS are also crucial regulators of prenatal and embryonic development (Dennery 2004) and immune system by triggering T cells proliferation through NF-κB activation (Tatla et al. 1999). However, during aging and age-related diseases, the machinery controlling ROS levels within the body become deregulated which in turn leads to oxidative stress at the later stages of life. Impairment of redox equilibrium causes both oxidative damage and the activation of pathways leading to chronic inflammation. During aging, ROS can induce ERK, JNK and p38 MAPK pathways and stimulate NF-κB-dependent transcription of pro-inflammatory cytokines (TNF-α, IL-1β, IL-6), chemokines (IL-8, RANTES), adhesion molecules (VCAM-1, ICAM-1, P- and E-selectin) and other pro-inflammatory enzymes like iNOS and COX-2 (Chung et al. 2006). The beneficial role of ROS in growth and development can be transformed to detrimental effects of oxidative stress at the later stages of life, in agreement with the antagonistic pleiotropy theory of aging (Williams 1957).
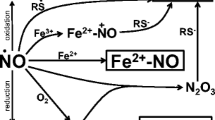
Recently, it was postulated that also NO• may mediate apoptotic cell death during CA in the yeast (Almeida et al. 2007). Nitric oxide level was reported to be elevated during CA in the yeast, as concluded from indirect measurements of NO• by Griess reaction (Almeida et al. 2007). It is believed that NO• may be considered as a signaling molecule also in unicellular organism like yeast controlling both apoptosis and sporulation (Kig and Temizkan 2009; Almeida et al. 2007). However, the endogenous source of NO• in the yeast cell remains a mystery due to the lack of mammalian nitric oxide synthase (NOS) orthologues in the yeast genome. Altogether, the role of NO• signaling during CA in the yeast requires further investigation.
Here, we used a variant of the standard procedure (Fabrizio and Longo 2003), similar to that used in the paper of Almeida et al. (2007) for measuring CLS of yeast lacking proteins involved in nitrosative stress responses and having imbalanced NO• production/level. We used the cells devoid of flavohemoglobin playing a major role in NO• detoxification, S-nitrosoglutathione reductase metabolizing S-nitrosoglutathione and the transcription factor Fzf1p mediating NO• response (Liu et al. 2000; Lewinska and Bartosz 2006; Lewinska et al. 2008; Sarver and DeRisi 2005; Foster et al. 2009). We measured the intracellular NO• level directly with a NO• specific probe (Balcerczyk et al. 2005) and checked the impact of medium (different glucose concentration) on NO• production during CA in the yeast. Our data suggest that NO• does not mediate apoptosis during CA of the yeast.
Materials and methods
Chemicals
DAF-FM diacetate and dihydroethidine were from Molecular Probes (Leiden, Netherlands). All other reagents, if not stated otherwise, were purchased from Sigma (Poznan, Poland) and were of analytical grade.
GSNO synthesis
0.5 M solution of GSH in 1 M HCl and 0.5 M solution of NaNO2 in distilled water were mixed on ice to obtain ~250 mM GSNO solution. The concentration of GSNO was measured spectrophotometrically at 332 nm (ε332 = 750 M−1 cm−1) (Hart 1985).
Strains and growth conditions
The following yeast strains were used:
-
(1)
Wild type strain BY4741 MATa his3 leu2 met15 ura3 and its isogenic mutants: Δyhb1, YGR234w::kanMX4; Δfzf1, YGL254w::kanMX4; Δsfa1, YDL168w::kanMX4 (Euroscarf, Germany)
-
(2)
A YHB1-GFP clone expressing full-length YHB1 gene containing an Aequorea victoria GFP (S56T) tag and HIS3 marker gene at the C-terminus in parental ATCC201388 MATa his3 leu2 met15 ura3 background (Huh et al. 2003) purchased from Invitrogen (Carlsbad, USA). The Yhb1-GFP fusion protein is integrated into the yeast chromosome through homologous recombination and is expressed using the endogenous promoter of YHB1 gene.
Yeast from one single colony was grown overnight in YPD medium (1% Difco Yeast Extract, 1% Difco Yeast Bacto-Peptone, 2% dextrose), washed and transferred into liquid YNB minimal medium (0.67% yeast nitrogen base with ammonium sulfate and without amino acids, 2% dextrose and with the essential amino acids supplementation) and cultured with shaking in a water bath incubator at 28°C for up to 2 weeks. Maximum population density is reached after the first 48 h of growth in YNB medium (approximately 4–5 × 107 cells/ml) and thus the number of colony forming units at day 2 (assuming that the dilution day is day 0) is considered as the initial survival (100% survival). At the indicated time points, appropriate aliquots were taken for analysis. Calorie restriction was achieved by using YNB medium with 0.5% dextrose.
YHB1-GFP strain and the disruptants were checked on YNB plates +Ura +Met +Leu −His, and on YPD Petri dishes containing 200 μg/ml geneticin (G418) sulphate, respectively.
Chronological life span (CLS) assays
For quantitative measurement of survival, the CFU assay and the kinetics of growth assay were used. For semiquantitative measurement of survival, the spot assay was used.
For the CFU assay, at the indicated time points (day 2, 4, 7 and 14), cells were removed, washed, diluted and spread onto YPD agar plates, and after 48 h CFUs/ml were counted (Fabrizio and Longo 2003).
For the kinetics of growth assay (Murakami and Kaeberlein 2009), at the indicated time points (day 2, 4, 7 and 14), cells were removed, washed, diluted, suspended in YPD medium (a total volume of 150 μl with working solution of 5 × 106 cells/ml) and put in a Heidolph Inkubator1000 at 1,200 rpm at 28°C. Their growth was monitored turbidimetrically at 600 nm in an Anthos 2010 type 17550 microplate reader every 2 h during a 12 h.
For the spot assay, at the indicated time points (day 2, 4, 7 and 14), cells were removed, washed, diluted (107, 106, 105, 104, 103 cells/ml in a volume of 2 μl), inoculated on solid YPD medium and inspected after 48 h.
Dead cells were estimated by methylene blue staining. At the indicated time points (day 2, 4, 7 and 14), the cells were washed with PBS, added to 100 μg/ml methylene blue in 2% sodium citrate dihydrate in a volume ratio of 1:1 and a total of 1,000 cells per each sample triplicate were analyzed in an optical microscope.
Apoptosis
After cell fixation with formaldehyde and zymolyase 100T treatment, evaluation of apoptotic-like changes (DNA fragmentation) in cells was performed with TUNEL (terminal dUTP nick-end labeling) assay with the in situ cell death detection kit with FITC-labeled nucleotides (Roche Diagnostics GmbH) using the standard protocol according to the manufacturer’s instructions. For each batch, a negative control without the addition of enzyme solution and a positive control with DNase I treatment were always included to ensure the reproducibility of the assay. For DNA visualization, the slides were counterstained with a drop of the mounting medium with 4′,6′-diamino-2-phenylindole (DAPI II) counterstain and a total of 200 cells per each sample triplicate were analyzed in an Olympus BX61 fluorescence microscope equipped with a DP72 CCD camera and Olympus CellF software.
Superoxide production
Cells were sedimented by centrifugation, washed twice and suspended in 0.1 M phosphate buffer (pH 7.0) containing 0.1% w/v glucose and 1 mM EDTA to a density of 108 cells/ml. Generation of superoxide was assessed with dihydroethidine (18.9 μM). Fluorescence intensity due to oxidation of dihydroethidine to ethidine was monitored in a Hitachi F2500 fluorescence spectrophotometer. Measurement conditions were: λex = 518 nm, λem = 605 nm; temperature 28°C.
Nitric oxide level
Cells were washed twice and suspended in 0.1 M phosphate buffer (pH 7.0) containing 1 mM EDTA and 0.1% glucose to a density of 108 cells/ml. Generation of intracellular nitric oxide was assessed with DAF-FM diacetate (1.1 μM). Fluorescence intensity was monitored in a Hitachi F2500 fluorescence spectrophotometer. Measurement conditions were: λex = 495 nm; λem = 515 nm; temperature 28°C. Additionally, flow cytometry analysis was performed. After given number of days, the cells were incubated with DAF-FM diacetate for 30 min and the median fluorescence intensity was measured in a Becton–Dickinson LSRII flow cytometer (San Jose, USA) equipped with a 488 nm argon laser. As a control, we used the cell suspension without DAF-FM diacetate to minimize the impact of the cell autofluorescence on the readings.
Yhb1p-GFP expression
The YHB1-GFP construct and wild type cells were grown in YPD medium to the late logarithmic phase. The cells were washed with 100 mM MES buffer, pH 6.5 and incubated in YNB medium for appropriate number of days. The median fluorescence intensity of the cells was measured in a Becton–Dickinson LSRII flow cytometer (San Jose, USA) equipped with a 488 nm argon laser. To exclude the autofluorescence effect, data standardization was performed. Autofluorescence of wild type strain was assumed as the constant value (1) and the median FITC-A results are the ratios of Yhb1p-GFP cells fluorescence and wild type cells fluorescence.
Products of the Griess reaction
Total NOx concentrations were measured in cell extracts according to Miranda et al. (Miranda et al. 2001). Briefly, a sample (4 volumes), VCl3 (8 mg/ml in 1 M HCl; 4 volumes), 0.1% N-(1-naphthyl)ethylenediamine dihydrochloride, NEDD; 2 volumes) and 2% sulfanilamide (SULF, in 5% HCl; 2 volumes) were mixed and incubated at 37°C for 30 min. The absorbance of the mixture was measured in a Cary Bio50 Varian spectrophotometer at 540 nm. Calculation was made on the basis of a standard curve obtained for a nitrate solution.
Protein tyrosine nitration
For dot blot, yeast protein extract (1 mg/ml) was directly blotted onto Immobilon-P membrane (Millipore Corporation, USA) and incubated with monoclonal anti-3-nitrotyrosine clone 18G4 (1:4,000 in 5% BSA in TBST, 1 h incubation) and then with anti-mouse IgG peroxidase conjugate (1:16,000 in 5% BSA in TBST, 1 h incubation). The dots were revealed on Hyperfilm ECL with ECL + detection kit (GE Healthcare). Images were analyzed with the NIH ImageJ software (v. 1.43) http://rsbweb.nih.gov/ij/ using the integrated density function. Background correction was performed using the subtract background command with the rolling ball radius set to 50 pixels. Rolling ball algorithm was inspired by the Stanley Sternberg’s article (Sternberg 1983). We evaluated the integrated density which is the sum of the values of the pixels of marked area of the image and is the equivalent to the product of area and mean gray value. The integrated fluorescence density is presented in relative units. For positive control, treatment with 5 mM S-nitrosoglutathione (GSNO) was used.
Statistical analysis
The results represent mean ± SD from at least three independent experiments. The obtained data conform the ANOVA assumptions as evaluated using Shapiro–Wilk normality test and Levene test for the equality of variances. Statistical significance of differences during CA in (i) the growth rate, (ii) superoxide, nitric oxide, products of Griess reaction generation and protein tyrosine nitration level, (iii) TUNEL positive cells and dead cells between wild type cells versus the Δyhb1, Δfzf1, and Δsfa1 cells, (iiii) in Yhb1p-GFP level and differences during CA concerning various dextrose concentration applied was evaluated using one-way analysis of variance (ANOVA) with post-hoc testing using a Dunnett’s multiple comparison test. P-values of less than 0.05 were considered significant. Statistical analysis of the data was performed using a StatSoft, Inc. (2005), STATISTICA, v. 7.0, http://www.statsoft.com/.
Results
We characterized the disruptants used (Δyhb1, Δfzf1, and Δsfa1) by the standard protocol used for CLS determination in the yeast (synthetic minimal medium with 2% dextrose and essential amino acids, flask culture with vigorous shaking). We employed the clonogenic assay i. e., the ability to form colonies on YPD agar plates, for estimation of cell survival (Fig. 1).
CLS of wild type strain and the isogenic Δyhb1, Δfzf1, and Δsfa1 mutants. After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis. The cells were spread onto YPD plates and the ability to form colonies was checked after 48 h. Wild type strain subjected to calorie restricted (CR) conditions is also shown. Bars indicate SD, n = 3; ***P < 0.001 as compared to wild type cells grown in standard growth conditions (one-way ANOVA and Dunnett’s multiple comparison test)
We were unable to record any differences in the percentage of survival between wild type strain and the mutants devoid of factors requested for proper nitrosative stress responses growing on standard CA medium (YNB, 2% dextrose). The survival was dramatically reduced at day 4 of culture and at day 7 only about 2% of cells were able to form colonies. In agreement with findings of other authors, calorie restricted (CR) conditions (YNB, 0.5% dextrose) extended CLS of all strains examined. The pattern of the extension of CLS was the same for all strains used (Fig. 1, only wild type is shown). Similar results were obtained for the spot assay and the kinetics of growth assay (Figs. 2 and 3).
Survival of a wild type strain and the isogenic Δyhb1, Δfzf1, and Δsfa1 mutants and b dextrose-dependent survival of wild type strain during CA (spot assay). After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis. Several dilutions (107, 106, 105, 104, 103 cells/ml) of a yeast CA culture in a volume of 2 μl were used, inoculated on solid YPD medium and inspected after 48 h. The results shown are representative for at least three independent experiments. CR calorie restriction, NR standard growth conditions
Kinetics of growth of chronologically aged cells: a wild type strain, calorie restriction (CR); b wild type strain, standard growth conditions (NR); c Δyhb1 (NR); d Δfzf1 (NR); e Δsfa1 (NR). After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis. A total volume of 150 μl YPD medium with working solution of 5 × 106 cells/ml were cultured at 28°C and their growth was monitored turbidimetrically at 600 nm in a microplate reader every 2 h during a 12 h. Bars indicate SD, n = 3
With the drop test, reflecting semiquantitatively the cell growth recovery, we showed that at day 7 of culture, the cells evidently almost lost their ability to divide (Fig. 2). The growth rate was also inhibited at day 4 of culture (up to 12 h of the growth) (Fig. 3).
Additionally, at day 4 almost a half of cell population was dead as estimated with methylene blue staining (Fig. 4).
The number of dead cells detected by methylene blue staining. After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis and a total of 1,000 stained cells per each sample triplicate were analyzed in an optical microscope. Mean values ± SD, n = 3; *P < 0.05, ***P < 0.001 as compared to wild type cells grown in standard growth conditions (one-way ANOVA and Dunnett’s multiple comparison test)
We also recorded unequivocal differences in the level of dead cells when the cells were cultured in the various conditions (different dextrose concentrations) (Fig. 4). CR was responsible for the same decrease in dead cell level in all strains examined (Fig. 4, only wild type is shown). We also checked the level of the cell DNA fragmentation which is considered as a one of the markers of apoptosis. At day 4, almost a half of cell population was apoptotic and at day 7 we were unable to record apoptosis (Fig. 5).
DNA fragmentation as a marker of apoptotic cell death during CA (TUNEL assay). After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis and a total of 200 cells per each sample triplicate were analyzed in a fluorescence microscope. Mean values ± SD, n = 3; *P < 0.05 as compared to wild type cells grown in standard growth conditions (one-way ANOVA and Dunnett’s multiple comparison test). In the case when apoptotic type of death could not be detected (high mortality, probably with high level of necrosis), it was denoted on the graph as “nd”
By then, probably nearly the whole population was dead which is in an agreement with data obtained by methylene blue staining (Fig. 4). CR yeast cultures exhibited reduced apoptosis occurrence compared to the standard growth conditions (Fig. 5).
Since free radicals and oxidative damage are thought to be implicated in the aging process (Harman 1956), we analyzed superoxide production during CA in the yeast (Fig. 6).
Intracellular superoxide production during CA estimated by the rate of increase of ethidine fluorescence. After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis and yeast cells were incubated with the fluorogenic probe, dihydroethidine in 0.1 M phosphate buffer (pH 7.0) containing 1 mM EDTA and 0.1% glucose at 28°C for 20 min. Mean values ± SD, n = 3; *P < 0.05, **P < 0.01, ***P < 0.001 as compared to wild type cells grown in standard growth conditions (one-way ANOVA and Dunnett’s multiple comparison test)
The cells with an insufficient protection against nitric oxide were not found to be more prone to oxidative stress during CA compared to wild type cells (Fig. 6) with the exception of the Δsfa1 strain which may be due to the fact that Sfa1p is both S-nitrosoglutathione reductase and alcohol dehydrogenase. However, only a 20–40% (statistically significant) increase was seen in the superoxide level in the Δsfa1 disruptant compared to the wild type strain during CA. Modulation of superoxide production by CR conditions (a two-fold decrease) was limited only to day 2 of culture (Fig. 6).
Finally, we tested the possibility that nitric oxide is elevated during CA in the yeast and nitric oxide is a signaling molecule triggering apoptosis during CA in the yeast as it was suggested by other authors on the basis of indirect measurements of nitric oxide by the Griess reaction (Almeida et al. 2007). We performed a direct analysis of intracellular nitric oxide production with a fluorogenic probe DAF-FM DA, fairly specific for nitric oxide. We found that nitric oxide is rather decreased than increased during CA in the yeast (Fig. 7). Under CR conditions, the basal level of nitric oxide was significantly higher than under standard growth conditions, but during CA its level also decreased (Fig. 7).
Generation of intracellular nitric oxide during CA assessed with DAF-FM DA. After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis and yeast cells were incubated with the fluorogenic probe, DAF-FM DA in 0.1 M phosphate buffer (pH 7.0) containing 1 mM EDTA and 0.1% glucose at 28°C for 30 min. Mean values ± SD, n = 3; ***P < 0.001 as compared to wild type cells grown in standard growth conditions (one-way ANOVA and Dunnett’s multiple comparison test)
Additionally, the Δyhb1, Δfzf1, and Δsfa1 mutants did not show higher levels of nitric oxide than wild type strain (Fig. 7). The results obtained fluorometrically were confirmed by flow cytometry (data not shown). We also employed the Griess reaction which provides an indirect estimation of nitric oxide level but in fact it reflects general nitrogen metabolism (total NOx in an analyzed sample). The products of Griess reaction dropped during CA in the yeast (Fig. 8).
We were unable to show any differences in the level of the Griess reaction products between strains and dextrose concentrations used (Fig. 8).
Altogether, our data showed the apoptosis occurs during CA in the yeast but is due to superoxide signaling rather than nitric oxide signaling.
Moreover, we checked if the cells with improper nitric oxide detoxification/signaling (the Δyhb1, Δfzf1, and Δsfa1 mutants) may be susceptible to nitrosative stress during CA. We analyzed protein tyrosine nitration which is mediated by secondary products of nitric oxide metabolism in the presence of oxidants. We showed no significant differences in protein tyrosine nitration between the wild type strain and the disruptants used (Fig. 9).
The level of protein tyrosine nitration during CA (a). After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis. Bars indicate SD, n = 3; *P < 0.05 as compared to wild type cells grown in standard growth conditions (one-way ANOVA and Dunnett’s multiple comparison test). The positive control for protein tyrosine nitration with 5 mM GSNO (b). Bars indicate SD, n = 3; **P < 0.01 as compared to wild type cells treated with 5 mM GSNO (one-way ANOVA and Dunnett’s multiple comparison test)
We also tested the importance of yeast flavohemoglobin, playing a pivotal role in nitric oxide detoxification, during CA in the yeast. Yhb1p level was significantly decreased, independently of dextrose concentration used, during CA in the yeast (Fig. 10).
Yhb1p expression during CA. After 2, 4, 7 and 14 days, appropriate aliquots from CA cultures were taken for analysis. The YHB1-GFP construct and wild type cells were grown in YPD medium to late logarithmic phase. The YHB1-GFP construct and wild type cells were washed with 100 mM MES buffer, pH 6.5 and the median fluorescence intensity of the cells was measured in a flow cytometer. Autofluorescence of the wild type strain was assumed as 1 and the median FITC-A results are the ratios of Yhb1p-GFP cells fluorescence and wild type cells fluorescence. Mean values ± SD, n = 3; ***P < 0.001 as compared to day 1 standard growth conditions and +++ P < 0.001 as compared to day one calorie restricted conditions (one-way ANOVA and Dunnett’s multiple comparison test)
Discussion
Nitric oxide at low concentrations is a very important signaling molecule in mammals involved in the regulation of vasodilation, neurotransmission, and penile erection (Moncada et al. 1991; Ignarro 2002). At much higher concentrations, attainable due to the action of an inducible nitric oxide synthase (iNOS), nitric oxide is toxic to the cells, and is used as a powerful weapon against pathogens (Nathan and Hibbs 1991; Tuteja et al. 2004).
It was postulated that nitric oxide may also act as a signaling molecule in unicellular eukaryotes like the yeast (Kig and Temizkan 2009; Almeida et al. 2007; Osorio et al. 2007). However, the exact source of nitric oxide within the yeast cell is unknown. Despite the fact that NOS-like activity was reported both in budding and fission yeast extracts (Osorio et al. 2007; Kig and Temizkan 2009), no true mammalian NOS orthologues were found in the yeast genome. Regardless of the proteins with cross-reactivity with the same antibodies against mammalian NOSs used were reported in budding yeast (Kanadia et al. 1998; Domitrovic et al. 2003), the visualized immunoreactive bands were of different molecular sizes. It was shown that NOS-like activity is required for sporulation during meiosis in fission yeast (Kig and Temizkan 2009) and controls hydrogen peroxide-induced apoptosis in budding yeast (Almeida et al. 2007). Additionally, nitric oxide signaling was claimed to be implicated in the programmed cell death during CA of the yeast, basing on indirect measurements of nitric oxide by the Griess reaction (Almeida et al. 2007). However, the assay was done in the supernatant of yeast cultures which may suggest that during CA the products of nitrogen metabolism are accumulated in the medium and an increased intracellular production of nitric oxide does not necessarily occurs.
Another possible source of nitric oxide in the yeast is the mitochondrial reduction of nitrite by cytochrome c oxidase, influenced by flavohemoglobin (Castello et al. 2006). However, this reaction, being a part of hypoxic signaling, can proceed only under hypoxic and anoxic conditions (Castello et al. 2006) and therefore seems to be not pertinent to normoxic conditions of CA methodology.
Since very little is known about the role of nitric oxide as a signaling molecule during CA in the yeast and nitric oxide is mainly considered as a nitrosative stress agent in this species, we decided to study the relationships between flavohemoglobin, nitric oxide signaling, apoptosis and aging in a more detailed.
We chose three yeast mutants lacking proteins known for their involvement in nitrosative stress responses (Liu et al. 2000; Lewinska and Bartosz 2006; Lewinska et al. 2008; Sarver and DeRisi 2005; Foster et al. 2009) and conducted chronological lifespan analysis with standard methodology and a direct monitoring of intracellular nitric oxide production with a nitric oxide specific probe (Balcerczyk et al. 2005). We recorded strain-independent decrease rather than increase in nitric oxide production during CA, a result being reproduced by a different methodology. Simultaneously, we showed an elevation of apoptosis during CA in the yeast which was evidently nitric oxide-independent. These data suggest that apoptosis during CA in the yeast is not mediated by nitric oxide. Nota bene, under CR conditions, nitric oxide production was significantly increased and in parallel apoptosis occurrence was reduced, compared to standard conditions of culture. Apparently, the link between CR and nitric oxide signaling requires further attention.
Moreover, yeast cells did not suffer nitrosative stress during CA since the pattern of protein tyrosine nitration was slightly decreased. Additionally, protection against nitrosative stress also seems not to be required during CA since (i) the CLS of the mutant cells with imbalanced pathways metabolizing nitric oxide and its secondary metabolic products was unaffected compared to wild type strain CLS and (ii) flavohemoglobin expression was dextrose concentration-independent and rather decreased than increased during CA.
According to the free radical theory of aging (Harman 1956), superoxide is thought to be a pro-aging factor accumulating during CA in the yeast and causing oxidative damage to macromolecules. However, CLS extension may be accompanied by an increase in oxidative damage (Mesquita et al. 2010) which may suggest that the pro-aging effects of oxidative stress is not necessary a direct outcome of oxidative damage in the yeast. Moreover, superoxide was found to be a signaling molecule modulating programmed cell death during CA in the yeast (Fabrizio et al. 2004). In this study, we found an elevation in superoxide level during CA, which was more clearly seen for standard growth conditions (2% dextrose in YNB medium) than under CR conditions (0.5% dextrose in YNB medium). It was shown that high glucose activates growth signaling which in turn causes an augmentation in superoxide production, inhibition in the stationary phase G0/G1 arrest, DNA replication stress, apoptosis and CLS shortening (Weinberger et al. 2010). It is thought that a similar mechanism may account for aging and age-related diseases linked to hyperglycemia and excess calorie intake like diabetes and cardiovascular diseases in higher eukaryotes (Weinberger et al. 2010). In mammals, high levels of glucose may stimulate AKT-dependent growth signaling, induce DNA damage, an increase in superoxide level and senescence (Sheu et al. 2004; Ksiazek et al. 2008; Yokoi et al. 2006; Lee and Han 2009).
Nevertheless, the proposed superoxide signaling during CA needs to be clarified in details especially the link between growth pathways and the generation of superoxide (specific/unspecific).
Taken together, our results argue against the regulatory role of nitric oxide during CA in the yeast.
References
Almeida B, Buttner S, Ohlmeier S, Silva A, Mesquita A, Sampaio-Marques B, Osorio NS, Kollau A, Mayer B, Leao C, Laranjinha J, Rodrigues F, Madeo F, Ludovico P (2007) NO-mediated apoptosis in yeast. J Cell Sci 120(Pt 18):3279–3288
Balcerczyk A, Soszynski M, Bartosz G (2005) On the specificity of 4-amino-5-methylamino-2′, 7′-difluorofluorescein as a probe for nitric oxide. Free Radic Biol Med 39(3):327–335
Bartosz G (2009) Reactive oxygen species: destroyers or messengers? Biochem Pharmacol 77(8):1303–1315
Berner YN, Stern F (2004) Energy restriction controls aging through neuroendocrine signal transduction. Ageing Res Rev 3(2):189–198
Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L (2010) Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11(1):35–46
Budovsky A, Abramovich A, Cohen R, Chalifa-Caspi V, Fraifeld V (2007) Longevity network: construction and implications. Mech Ageing Dev 128(1):117–124
Campisi J (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120(4):513–522
Castello PR, David PS, McClure T, Crook Z, Poyton RO (2006) Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab 3(4):277–287
Chen Q, Ding Q, Keller JN (2005) The stationary phase model of aging in yeast for the study of oxidative stress and age-related neurodegeneration. Biogerontology 6(1):1–13
Chiarugi P, Cirri P (2003) Redox regulation of protein tyrosine phosphatases during receptor tyrosine kinase signal transduction. Trends Biochem Sci 28(9):509–514
Chung HY, Sung B, Jung KJ, Zou Y, Yu BP (2006) The molecular inflammatory process in aging. Antioxid Redox Signal 8(3–4):572–581
Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A (2006) Opposing activities protect against age-onset proteotoxicity. Science 313(5793):1604–1610
de Magalhaes JP, Church GM (2006) Cells discover fire: employing reactive oxygen species in development and consequences for aging. Exp Gerontol 41(1):1–10
de Magalhaes JP, Budovsky A, Lehmann G, Costa J, Li Y, Fraifeld V, Church GM (2009) The human ageing genomic resources: online databases and tools for biogerontologists. Aging Cell 8(1):65–72
Dennery PA (2004) Role of redox in fetal development and neonatal diseases. Antioxid Redox Signal 6(1):147–153
Dolle ME, Vijg J (2002) Genome dynamics in aging mice. Genome Res 12(11):1732–1738
Domitrovic T, Palhano FL, Barja-Fidalgo C, DeFreitas M, Orlando MT, Fernandes PM (2003) Role of nitric oxide in the response of Saccharomyces cerevisiae cells to heat shock and high hydrostatic pressure. FEMS Yeast Res 3(4):341–346
Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Frohlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F (2009) Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11(11):1305–1314
Esposito F, Ammendola R, Faraonio R, Russo T, Cimino F (2004) Redox control of signal transduction, gene expression and cellular senescence. Neurochem Res 29(3):617–628
Fabrizio P, Longo VD (2003) The chronological life span of Saccharomyces cerevisiae. Aging Cell 2(2):73–81
Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292(5515):288–290
Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD (2004) Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol 166(7):1055–1067
Foster MW, Liu L, Zeng M, Hess DT, Stamler JS (2009) A genetic analysis of nitrosative stress. Biochemistry 48(4):792–799
Gershon H, Gershon D (2000) The budding yeast, Saccharomyces cerevisiae, as a model for aging research: a critical review. Mech Ageing Dev 120(1–3):1–22
Harley CB, Vaziri H, Counter CM, Allsopp RC (1992) The telomere hypothesis of cellular aging. Exp Gerontol 27(4):375–382
Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11(3):298–300
Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460(7253):392–395
Hart TW (1985) Some observations concerning the S-nitroso and S-phenylsulphonyl derivatives of l-cysteine and glutathione. Tetrahedron Lett 26:2013–2016
Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F (2004) Chronological aging leads to apoptosis in yeast. J Cell Biol 164(4):501–507
Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686–691
Ignarro LJ (2002) Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J Physiol Pharmacol 53(4 Pt 1):503–514
Jakubowski W, Bilinski T, Bartosz G (2000) Oxidative stress during aging of stationary cultures of the yeast Saccharomyces cerevisiae. Free Radic Biol Med 28(5):659–664
Kaeberlein M (2010) Lessons on longevity from budding yeast. Nature 464(7288):513–519
Kanadia RN, Kuo WN, McNabb M, Botchway A (1998) Constitutive nitric oxide synthase in Saccharomyces cerevisiae. Biochem Mol Biol Int 45(6):1081–1087
Kennedy BK, Steffen KK, Kaeberlein M (2007) Ruminations on dietary restriction and aging. Cell Mol Life Sci 64(11):1323–1328
Kig C, Temizkan G (2009) Nitric oxide as a signaling molecule in the fission yeast Schizosaccharomyces pombe. Protoplasma 238(1–4):59–66
Ksiazek K, Passos JF, Olijslagers S, von Zglinicki T (2008) Mitochondrial dysfunction is a possible cause of accelerated senescence of mesothelial cells exposed to high glucose. Biochem Biophys Res Commun 366(3):793–799
Lee YJ, Han HJ (2009) Troglitazone ameliorates high glucose-induced EMT and dysfunction of SGLTs through PI3 K/Akt, GSK-3{beta}, Snail1, and {beta}-catenin in renal proximal tubule cells. Am J Physiol Renal Physiol. doi:10.1152/ajprenal.00475.2009
Lewinska A, Bartosz G (2006) Yeast flavohemoglobin protects against nitrosative stress and controls ferric reductase activity. Redox Rep 11(5):231–239
Lewinska A, Grzelak A, Bartosz G (2008) Application of a YHB1-GFP reporter to detect nitrosative stress in yeast. Redox Rep 13(4):161–171
Liu L, Zeng M, Hausladen A, Heitman J, Stamler JS (2000) Protection from nitrosative stress by yeast flavohemoglobin. Proc Natl Acad Sci USA 97(9):4672–4676
Longo VD, Gralla EB, Valentine JS (1996) Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem 271(21):12275–12280
MacLean M, Harris N, Piper PW (2001) Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of post-mitotic tissues in higher organisms. Yeast 18(6):499–509
Mesquita A, Weinberger M, Silva A, Sampaio-Marques B, Almeida B, Leao C, Costa V, Rodrigues F, Burhans WC, Ludovico P (2010) Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci USA 107(34):15123–15128
Miranda KM, Espey MG, Wink DA (2001) A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5(1):62–71
Moncada S, Palmer RM, Higgs EA (1991) Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev 43(2):109–142
Mortimer RK, Johnston JR (1959) Life span of individual yeast cells. Nature 183(4677):1751–1752
Murakami C, Kaeberlein M (2009) Quantifying yeast chronological life span by outgrowth of aged cells. J Vis Exp 6(27). doi:10.3791/1156
Nathan CF, Hibbs JB Jr (1991) Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr Opin Immunol 3(1):65–70
Osorio NS, Carvalho A, Almeida AJ, Padilla-Lopez S, Leao C, Laranjinha J, Ludovico P, Pearce DA, Rodrigues F (2007) Nitric oxide signaling is disrupted in the yeast model for Batten disease. Mol Biol Cell 18(7):2755–2767
Powers RW III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S (2006) Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev 20(2):174–184
Remacle J, Raes M, Toussaint O, Renard P, Rao G (1995) Low levels of reactive oxygen species as modulators of cell function. Mutat Res 316(3):103–122
Sarver A, DeRisi J (2005) Fzf1p regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol Biol Cell 16(10):4781–4791
Sheu ML, Ho FM, Chao KF, Kuo ML, Liu SH (2004) Activation of phosphoinositide 3-kinase in response to high glucose leads to regulation of reactive oxygen species-related nuclear factor-kappaB activation and cyclooxygenase-2 expression in mesangial cells. Mol Pharmacol 66(1):187–196
Sternberg S (1983) Biomedical image processing. Computer 16(1):22–34
Tatla S, Woodhead V, Foreman JC, Chain BM (1999) The role of reactive oxygen species in triggering proliferation and IL-2 secretion in T cells. Free Radic Biol Med 26(1–2):14–24
Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW (2004) Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci USA 101(17):6564–6569
Tuteja N, Chandra M, Tuteja R, Misra MK (2004) Nitric oxide as a unique bioactive signaling messenger in physiology and pathophysiology. J Biomed Biotechnol 2004(4):227–237
Weinberger M, Mesquita A, Caroll T, Marks L, Yang H, Zhang Z, Ludovico P, Burhans WC (2010) Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany NY) 2(10):709–726
Williams G (1957) Pleiotropy, natural selection, and the evolution of senescence. Evolution 11:398–411
Yan L, Vatner DE, O’Connor JP, Ivessa A, Ge H, Chen W, Hirotani S, Ishikawa Y, Sadoshima J, Vatner SF (2007) Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell 130(2):247–258
Yokoi T, Fukuo K, Yasuda O, Hotta M, Miyazaki J, Takemura Y, Kawamoto H, Ichijo H, Ogihara T (2006) Apoptosis signal-regulating kinase 1 mediates cellular senescence induced by high glucose in endothelial cells. Diabetes 55(6):1660–1665
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lewinska, A., Macierzynska, E., Grzelak, A. et al. A genetic analysis of nitric oxide-mediated signaling during chronological aging in the yeast. Biogerontology 12, 309–320 (2011). https://doi.org/10.1007/s10522-011-9329-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10522-011-9329-4