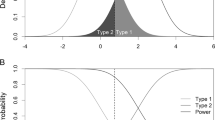
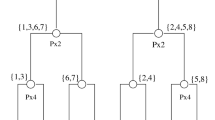
Abstract
Knowledge about the proportion of markers without effects (p 0 ) and the effect sizes in large scale genetic studies is important to understand the basic properties of the data and for applications such as the control of false discoveries and designing adequately powered replication studies. Many p 0 estimators have been proposed. However, high dimensional data sets typically comprise a large range of effect sizes and it is unclear whether the estimated p 0 is related to the whole range, including markers with very small effects, or just the markers with large effects. In this article we develop an estimation procedure that can be used in all scenarios where the test statistic distribution under the alternative can be characterized by a single parameter (e.g. non-centrality parameter of the non-central chi-square or F distribution). The estimation procedure starts with estimating the largest effect in the data set, then the second largest effect, then the third largest effect, etc. We stop when the effect sizes become so small that they cannot be estimated precisely anymore for the given sample size. Once the individual effect sizes are estimated, they can be used to calculate an interpretable estimate of p 0. Thus, our method results in both an interpretable estimate of p 0 as well as estimates of the effect sizes present in the whole marker set by repeatedly estimating a single parameter. Simulations suggest that the effects are estimated precisely with only a small upward bias. The R codes that compute the effect estimates are freely downloadable from the website: http://www.people.vcu.edu/~jbukszar/.



Similar content being viewed by others
References
Agresti A (1990) Categorical data analysis. New York
Allison DB, Gadbury G, Heo M, Fernandez J, Lee C-K, Prolla TA, Weindruch R (2002) A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data Anal 39:1–20
Benjamini Y, Hochberg Y (2000) On adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat 25:60–83
Bukszár J, Van den Oord EJCG (2005) Accurate and efficient power calculations for 2 × m tables in unmatched case-control designs. Stat Med 25:2632–2646
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74(1):106–120
Cohen J (1988) Statistical power analysis for the behavioral sciences. Erlbaum, Hillsdale
Dalmasso C, Broet P, Moreau T (2005) A simple procedure for estimating the false discovery rate. Bioinformatics 21:660–668
Delongchamp RR, Bowyer JF, Chen JJ, Kodell RL (2004) Multiple-testing strategy for analyzing cDNA array data on gene expression. Biometrics 60(3):774–782
Efron B, Tibshirani R, Storey JD, Tusher VG (2001) Empirical Bayes analysis of a microarray experiment. J Am Stat Assoc 96:1151–1160
Genovese C, Wasserman L (2002) Operating characteristics and extensions of the false discovery rate procedure. J R Stat Soc B 64:499–517
Genovese C, Wasserman L (2004) A stochastic process approach to false discovery control. Ann Stat 32:1035–1061
Ghosh A, Zou F, Wright FA (2008) Estimating odds ratios in genome scans: an approximate conditional likelihood approach. Am J Hum Genet 82(5):1064–1074
Goring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69(6):1357–1369
Hayes B, Goddard ME (2001) The distribution of the effects of genes affecting quantitative traits in livestock. Genet Sel Evol 33(3):209–229
Hsueh H, Chen J, Kodell R (2003) Comparison of methods for estimating the number of true null hypotheses in multiplicity testing. J Biopharm Stat 13:675–689
Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG (2001) Replication validity of genetic association studies. Nat Genet 29(3):306–309
Kuo PH, Bukszar J, van den Oord EJ (2007) Estimating the number and size of the main effects in genome-wide case-control association studies. BMC Proc 1(Suppl 1):S143
Meinshausen N, Rice J (2006) Estimating the proportion of false null hypotheses among a large number of independently tested hypotheses. Ann Stat 34(1):373–393
Mosig MO, Lipkin E, Khutoreskaya G, Tchourzyna E, Soller M, Friedmann A (2001) A whole genome scan for quantitative trait loci affecting milk protein percentage in Israeli-Holstein cattle, by means of selective milk DNA pooling in a daughter design, using an adjusted false discovery rate criterion. Genetics 157(4):1683–1698
Pounds S, Cheng C (2004) Improving false discovery rate estimation. Bioinformatics 20(11):1737–1745
Pounds S, Morris SW (2003) Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 19(10):1236–1242
Sarkar S (2002) Some results on false discovery rate in stepwise multiple testing procedures. Ann Stat 30:239–257
Sarkar S (2004) FDR-controlling stepwise procedures and their false negative rates. J Stat Plan Inference 125:119–137
Schweder T, Spjøtvoll E (1982) Plots of p-values to evaluate many tests simultaneously. Biometrika 69:493–502
Storey J (2002) A direct approach to false discovery rates. J R Stat Soc B 64:479–498
Taylor J, Tibshirani R, Efron B (2005) The ‘miss rate’ for the analysis of gene expression data. Biostatistics 6(1):111–117
Turkheimer FE, Smith CB, Schmidt K (2001) Estimation of the number of “true” null hypotheses in multivariate analysis of neuroimaging data. Neuroimage 13(5):920–930
van den Oord EJ, Kuo PH, Hartmann AM, Webb BT, Moller HJ, Hettema JM, Giegling I, Bukszar J, Rujescu D (2008) Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatry 65(9):1062–1071
Weir BS (1996) Genetic data analysis II. Sunderland
Zhong H, Prentice RL (2008) Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics 9(4):621–634
Zollner S, Pritchard JK (2007) Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am J Hum Genet 80(4):605–615
Acknowledgments
This work was supported by grant R01HG004240.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Stacey Cherny.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bukszár, J., van den Oord, E.J.C.G. Estimating Effect Sizes in Genome-Wide Association Studies. Behav Genet 40, 394–403 (2010). https://doi.org/10.1007/s10519-009-9321-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-009-9321-9