Abstract
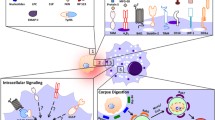
Apoptotic corpses can be engulfed and cleared by many other cell types in addition to ‘professional’ phagocytes such as macrophage. Studies of several organisms have contributed to the understanding of apoptotic corpse engulfment. Two partially redundant engulfment pathways have been characterized that act even in non-professional phagocytes to promote corpse engulfment. This review summarizes some recent progress in signaling by these pathways, including the exposure of eat-me-signals on apoptotic cells, and insights from Drosophila on the roles of the bridging receptor Six Microns Under, the non-receptor tyrosine kinase Shark, and store-operated calcium release in the Draper/Ced-1 pathway of corpse recognition and internalization. The mechanism of apoptotic phagosome maturation is outlined, and possible connections between corpse engulfment and proliferation, cell competition, and immunity are discussed.


Similar content being viewed by others
References
Krieser RJ, White K (2002) Engulfment mechanism of apoptotic cells. Curr Opin Cell Biol 14:734–738. doi:10.1016/S0955-0674(02)00390-3
Reddien PW, Horvitz HR (2004) The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol 20:193–221. doi:10.1146/annurev.cellbio.20.022003.114619
Mangahas PM, Zhou Z (2005) Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol 16:295–306. doi:10.1016/j.semcdb.2004.12.005
Erwig LP, Henson PM (2008) Clearance of apoptotic cells by phagocytes. Cell Death Differ 15:243–250. doi:10.1038/sj.cdd.4402184
Fristrom D (1969) Cellular degeneration in the production of some mutant phenotypes in Drosophila melanogaster. Mol Gen Genet 103:363–379. doi:10.1007/BF00383486
Franc NC, White K, Ezekowitz RA (1999) Phagocytosis and development: back to the future. Curr Opin Immunol 11:47–52. doi:10.1016/S0952-7915(99)80009-0
Crozatier M, Meister M (2007) Drosophila haematopoiesis. Cell Microbiol 9:1117–1126. doi:10.1111/j.1462-5822.2007.00930.x
Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA (1996) Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4:431–443. doi:10.1016/S1074-7613(00)80410-0
Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM (1992) Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148:2207–2216
Williamson P, Schlegel RA (2002) Transbilayer phospholipid movement and the clearance of apoptotic cells. Biochim Biophys Acta 1585:53–63
Bratton DL, Fadok VA, Richter DA, Kailey JM, Guthrie LA, Henson PM (1997) Appearance of phosphatidylserine on apoptotic cells requires calcium-mediated nonspecific flip-flop and is enhanced by loss of the aminophospholipid translocase. J Biol Chem 272:26159–26165. doi:10.1074/jbc.272.42.26159
Daleke DL, Lyles JV (2000) Identification and purification of aminophospholipid flippases. Biochim Biophys Acta 1486:108–127
Vance JE, Steenbergen R (2005) Metabolism and functions of phosphatidylserine. Prog Lipid Res 44:207–234. doi:10.1016/j.plipres.2005.05.001
Gardai SJ, Bratton DL, Ogden CA, Henson PM (2006) Recognition ligands on apoptotic cells: a perspective. J Leukoc Biol 79:896–903. doi:10.1189/jlb.1005550
van den Eijnde SM, Boshart L, Baehrecke EH, De Zeeuw CI, Reutelingsperger CP, Vermeij-Keers C (1998) Cell surface exposure of phosphatidylserine during apoptosis is phylogenetically conserved. Apoptosis 3:9–16. doi:10.1023/A:1009650917818
Zwaal RF, Comfurius P, Bevers EM (2005) Surface exposure of phosphatidylserine in pathological cells. Cell Mol Life Sci 62:971–988. doi:10.1007/s00018-005-4527-3
Wang X, Wang J, Gengyo-Ando K et al (2007) C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol 9:541–549. doi:10.1038/ncb1574
Venegas V, Zhou Z (2007) Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol Biol Cell 18:3180–3192. doi:10.1091/mbc.E07-02-0138
Modjtahedi N, Giordanetto F, Madeo F, Kroemer G (2006) Apoptosis-inducing factor: vital and lethal. Trends Cell Biol 16:264–272. doi:10.1016/j.tcb.2006.03.008
Susin SA, Lorenzo HK, Zamzami N et al (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441–446. doi:10.1038/17135
Halleck MS, Pradhan D, Blackman C, Berkes C, Williamson P, Schlegel RA (1998) Multiple members of a third subfamily of P-type ATPases identified by genomic sequences and ESTs. Genome Res 8:354–361
Ruaud AF, Nilsson L, Richard F, Larsen MK, Bessereau JL, Tuck S (2008) The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic 10:88–100
Zullig S, Neukomm LJ, Jovanovic M et al (2007) Aminophospholipid translocase TAT-1 promotes phosphatidylserine exposure during C. elegans apoptosis. Curr Biol 17:994–999. doi:10.1016/j.cub.2007.05.024
Darland-Ransom M, Wang X, Sun CL et al (2008) Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320:528–531. doi:10.1126/science.1155847
Kuraishi T, Manaka J, Kono M et al (2007) Identification of calreticulin as a marker for phagocytosis of apoptotic cells in Drosophila. Exp Cell Res 313:500–510. doi:10.1016/j.yexcr.2006.10.027
Gardai SJ, McPhillips KA, Frasch SC et al (2005) Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123:321–334. doi:10.1016/j.cell.2005.08.032
Park D, Tosello-Trampont AC, Elliott MR et al (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450:430–434. doi:10.1038/nature06329
Zhou Z, Hartwieg E, Horvitz HR (2001) CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104:43–56. doi:10.1016/S0092-8674(01)00190-8
Manaka J, Kuraishi T, Shiratsuchi A et al (2004) Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem 279:48466–48476. doi:10.1074/jbc.M408597200
Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ (2003) Unwrapping glial biology: gcm target genes regulating glial development, diversification, and function. Neuron 38:567–580. doi:10.1016/S0896-6273(03)00289-7
MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR (2006) The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50:869–881. doi:10.1016/j.neuron.2006.04.028
Li W, Baker NE (2007) Engulfment is required for cell competition. Cell 129:1215–1225. doi:10.1016/j.cell.2007.03.054
Cuttell L, Vaughan A, Silva E et al (2008) Undertaker, a Drosophila junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell 135:524–534. doi:10.1016/j.cell.2008.08.033
Su HP, Nakada-Tsukui K, Tosello-Trampont AC et al (2002) Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J Biol Chem 277:11772–11779. doi:10.1074/jbc.M109336200
Awasaki T, Tatsumi R, Takahashi K et al (2006) Essential role of the apoptotic cell engulfment genes Draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50:855–867. doi:10.1016/j.neuron.2006.04.027
Kurucz E, Markus R, Zsamboki J et al (2007) Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol 17:649–654. doi:10.1016/j.cub.2007.02.041
Kocks C, Cho JH, Nehme N et al (2005) Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123:335–346. doi:10.1016/j.cell.2005.08.034
Kurant E, Axelrod S, Leaman D, Gaul U (2008) Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133:498–509. doi:10.1016/j.cell.2008.02.052
Elliott MR, Ravichandran KS (2008) Death in the CNS: six-microns-under. Cell 133:393–395. doi:10.1016/j.cell.2008.04.014
Gumienny TL, Brugnera E, Tosello-Trampont AC et al (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107:27–41. doi:10.1016/S0092-8674(01)00520-7
Lu M, Kinchen JM, Rossman KL et al (2004) PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat Struct Mol Biol 11:756–762. doi:10.1038/nsmb800
Chimini G, Chavrier P (2000) Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nat Cell Biol 2:E191–E196. doi:10.1038/35036454
Ravichandran KS, Lorenz U (2007) Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol 7:964–974. doi:10.1038/nri2214
Albert ML, Kim JI, Birge RB (2000) Alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol 2:899–905. doi:10.1038/35046549
Kinchen JM, Cabello J, Klingele D et al (2005) Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 434:93–99. doi:10.1038/nature03263
Leverrier Y, Lorenzi R, Blundell MP et al (2001) Cutting edge: the Wiskott–Aldrich syndrome protein is required for efficient phagocytosis of apoptotic cells. J Immunol 166:4831–4834
Tsuboi S, Meerloo J (2007) Wiskott–Aldrich syndrome protein is a key regulator of the phagocytic cup formation in macrophages. J Biol Chem 282:34194–34203. doi:10.1074/jbc.M705999200
Pearson AM, Baksa K, Ramet M et al (2003) Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila. Microbes Infect 5:815–824. doi:10.1016/S1286-4579(03)00157-6
Yu X, Odera S, Chuang CH, Lu N, Zhou Z (2006) C. elegans dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell 10:743–757. doi:10.1016/j.devcel.2006.04.007
Rubartelli A, Poggi A, Zocchi MR (1997) The selective engulfment of apoptotic bodies by dendritic cells is mediated by the alpha(v)beta3 integrin and requires intracellular and extracellular calcium. Eur J Immunol 27:1893–1900. doi:10.1002/eji.1830270812
Dewitt S, Hallett MB (2002) Cytosolic free Ca(2+) changes and calpain activation are required for beta integrin-accelerated phagocytosis by human neutrophils. J Cell Biol 159:181–189. doi:10.1083/jcb.200206089
Dewitt S, Laffafian I, Hallett MB (2003) Phagosomal oxidative activity during beta2 integrin (CR3)-mediated phagocytosis by neutrophils is triggered by a non-restricted Ca2+ signal: Ca2+ controls time not space. J Cell Sci 116:2857–2865. doi:10.1242/jcs.00499
Tejle K, Magnusson KE, Rasmusson B (2002) Phagocytosis and phagosome maturation are regulated by calcium in J774 macrophages interacting with unopsonized prey. Biosci Rep 22:529–540. doi:10.1023/A:1022025903688
Roos J, DiGregorio PJ, Yeromin AV et al (2005) STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol 169:435–445. doi:10.1083/jcb.200502019
Zhang SL, Yu Y, Roos J et al (2005) STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437:902–905. doi:10.1038/nature04147
Vig M, Peinelt C, Beck A et al (2006) CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312:1220–1223. doi:10.1126/science.1127883
Feske S, Gwack Y, Prakriya M et al (2006) A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441:179–185. doi:10.1038/nature04702
Desjardins M (2003) ER-mediated phagocytosis: a new membrane for new functions. Nat Rev Immunol 3:280–291. doi:10.1038/nri1053
Touret N, Paroutis P, Terebiznik M et al (2005) Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell 123:157–170. doi:10.1016/j.cell.2005.08.018
Ziegenfuss JS, Biswas R, Avery MA et al (2008) Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature 453:935–939. doi:10.1038/nature06901
Fernandez R, Takahashi F, Liu Z, Steward R, Stein D, Stanley ER (2000) The Drosophila shark tyrosine kinase is required for embryonic dorsal closure. Genes Dev 14:604–614
Biswas R, Stein D, Stanley ER (2006) Drosophila dok is required for embryonic dorsal closure. Development 133:217–227. doi:10.1242/dev.02198
Wang X, Sada K, Yanagi S, Yang C, Rezaul K, Yamamura H (1994) Intracellular calcium dependent activation of p72syk in platelets. J Biochem 116:858–861
Papp S, Fadel MP, Kim H, McCulloch CA, Opas M (2007) Calreticulin affects fibronectin-based cell-substratum adhesion via the regulation of c-Src activity. J Biol Chem 282:16585–16598. doi:10.1074/jbc.M701011200
Feske S (2007) Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol 7:690–702. doi:10.1038/nri2152
Yu X, Lu N, Zhou Z (2008) Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol 6:e61. doi:10.1371/journal.pbio.0060061
Kinchen JM, Ravichandran KS (2008) Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol 9:781–795. doi:10.1038/nrm2515
Stuart LM, Boulais J, Charriere GM et al (2007) A systems biology analysis of the Drosophila phagosome. Nature 445:95–101. doi:10.1038/nature05380
Garin J, Diez R, Kieffer S et al (2001) The phagosome proteome: insight into phagosome functions. J Cell Biol 152:165–180. doi:10.1083/jcb.152.1.165
Vieira OV, Botelho RJ, Grinstein S (2002) Phagosome maturation: aging gracefully. Biochem J 366:689–704
Henry RM, Hoppe AD, Joshi N, Swanson JA (2004) The uniformity of phagosome maturation in macrophages. J Cell Biol 164:185–194. doi:10.1083/jcb.200307080
Desjardins M, Huber LA, Parton RG, Griffiths G (1994) Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol 124:677–688. doi:10.1083/jcb.124.5.677
Olkkonen VM, Stenmark H (1997) Role of Rab GTPases in membrane traffic. Int Rev Cytol 176:1–85. doi:10.1016/S0074-7696(08)61608-3
Kinchen JM, Doukoumetzidis K, Almendinger J et al (2008) A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol 10:556–566. doi:10.1038/ncb1718
Booth JW, Trimble WS, Grinstein S (2001) Membrane dynamics in phagocytosis. Semin Immunol 13:357–364. doi:10.1006/smim.2001.0332
Botelho RJ, Scott CC, Grinstein S (2004) Phosphoinositide involvement in phagocytosis and phagosome maturation. Curr Top Microbiol Immunol 282:1–30
Vieira OV, Bucci C, Harrison RE et al (2003) Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol 23:2501–2514. doi:10.1128/MCB.23.7.2501-2514.2003
Lu Q, Zhang Y, Hu T, Guo P, Li W, Wang X (2008) C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development 135:1069–1080. doi:10.1242/dev.016063
Mangahas PM, Yu X, Miller KG, Zhou Z (2008) The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol 180:357–373. doi:10.1083/jcb.200708130
Rink J, Ghigo E, Kalaidzidis Y, Zerial M (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122:735–749. doi:10.1016/j.cell.2005.06.043
Xiao H, Chen D, Fang Z et al (2009) Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell 20:21–32. doi:10.1091/mbc.E08-04-0441
Huynh KK, Grinstein S (2008) Phagocytosis: dynamin’s dual role in phagosome biogenesis. Curr Biol 18:R563–R565. doi:10.1016/j.cub.2008.05.032
Fan Y, Bergmann A (2008) Apoptosis-induced compensatory proliferation. The cell is dead. Long live the cell!. Trends Cell Biol 18:467–473. doi:10.1016/j.tcb.2008.08.001
Russell MA (1974) Pattern formation in the imaginal discs of a temperature-sensitive cell-lethal mutant of Drosophila melanogaster. Dev Biol 40:24–39. doi:10.1016/0012-1606(74)90104-3
Simpson P, Schneiderman HA (1975) Isolation of temperature-sensitive mutations blocking clone development in Drosophila melanogaster, and the effects of a temperature-sensitive cell lethal mutation on pattern formation in imaginal discs. Wilhelm Roux Arch Dev Biol 178:247–275. doi:10.1007/BF00848432
Hoeppner DJ, Hengartner MO, Schnabel R (2001) Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412:202–206. doi:10.1038/35084103
Reddien PW, Cameron S, Horvitz HR (2001) Phagocytosis promotes programmed cell death in C. elegans. Nature 412:198–202. doi:10.1038/35084096
Morata G, Ripoll P (1975) Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev Biol 42:211–221. doi:10.1016/0012-1606(75)90330-9
Marygold SJ, Roote J, Reuter G et al (2007) The ribosomal protein genes and minute loci of Drosophila melanogaster. Genome Biol 8:R216. doi:10.1186/gb-2007-8-10-r216
de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA (2004) Drosophila myc regulates organ size by inducing cell competition. Cell 117:107–116. doi:10.1016/S0092-8674(04)00214-4
Senoo-Matsuda N, Johnston LA (2007) Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc Natl Acad Sci USA 104:18543–18548. doi:10.1073/pnas.0709021104
Moreno E, Basler K (2004) dMyc transforms cells into super-competitors. Cell 117:117–129. doi:10.1016/S0092-8674(04)00262-4
Wang X, Wu YC, Fadok VA et al (2003) Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302:1563–1566. doi:10.1126/science.1087641
Kreiser R, Moore FE, Dresnek D et al (2007) The Drosophila homolog of the putative phosphatidylserine receptor functions to inhibit apoptosis. Development 134:2707–2714
Wu YC, Tibrewal N, Birge RB (2006) Phosphatidylserine recognition by phagocytosis: a view to kill. Trends Cell Biol 16:189–197. doi:10.1016/j.tcb.2006.02.003
Acknowledgments
We thank Diane Cox and Xiaochen Wang for helpful comments. Research in our laboratory is supported by grants from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. F. Fullard and A. Kale contributed equally.
Rights and permissions
About this article
Cite this article
Fullard, J.F., Kale, A. & Baker, N.E. Clearance of apoptotic corpses. Apoptosis 14, 1029–1037 (2009). https://doi.org/10.1007/s10495-009-0335-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-009-0335-9