Abstract
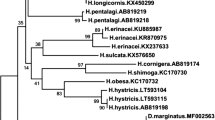
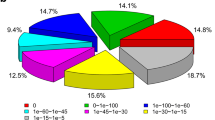
Ornithonyssus bacoti (Hirst) (Acari: Macronyssidae) is a vector and reservoir of pathogens causing serious infectious diseases, such as epidemic hemorrhagic fever, endemic typhus, tularemia, and leptospirosis. Its genome and transcriptome data are lacking in public databases. In this study, total RNA was extracted from live O. bacoti to conduct RNA-seq, functional annotation, coding domain sequence (CDS) prediction and simple sequence repeats (SSRs) detection. The results showed that 65.8 million clean reads were generated and assembled into 72,185 unigenes, of which 49.4% were annotated by seven functional databases. 23,121 unigenes were annotated and assigned to 457 species by non-redundant protein sequence database. The BLAST top-two hit species were Metaseiulus occidentalis and Ixodes scapularis. The procedure detected 12,426 SSRs, of which tri- and di-nucleotides were the most abundant types and the representative motifs were AAT/ATT and AC/GT. 26,936 CDS were predicted with a mean length of 711 bp. 87 unigenes of 30 functional genes, which are usually involved in stress responses, drug resistance, movement, metabolism and allergy, were further identified by bioinformatics methods. The unigenes putatively encoding cytochrome P450 proteins were further analyzed phylogenetically. In conclusion, this study completed the RNA-seq and functional annotation of O. bacoti successfully, which provides reliable molecular data for its future studies of gene function and molecular markers.









Similar content being viewed by others
References
Andersen JF, Hinnebusch BJ, Lucas DA, Conrads TP, Veenstra TD, Pham VM, Ribeiro JC (2007) An insight into the sialome of the oriental rat flea, Xenopsylla cheopis (Rots). BMC Genom 8(1):102–118. https://doi.org/10.1186/1471-2164-8-102
Bajda S, Dermauw W, Greenhalgh R, Nauen R, Tirry L, Clark RM, Van Leeuwen T (2015) Transcriptome profiling of a spirodiclofen susceptible and resistant strain of the European red mite Panonychus ulmi using strand-specific RNA-seq. BMC Genom 16(1):974–999. https://doi.org/10.1186/s12864-015-2157-1
Cabrera AR, Donohue KV, Khalil SMS, Scholl E, Opperman C, Sonenshine DE, Roe RM (2011) New approach for the study of mite reproduction: the first transcriptome analysis of a mite, Phytoseiulus persimilis (Acari: Phytoseiidae). J Insect Physiol 57(1):52–61. https://doi.org/10.1016/j.jinsphys.2010.09.006
Caragata EP, Pais FS, Baton LA, Silva JBL, Sorgine MHF, Moreira LA (2017) The transcriptome of the mosquito Aedes fluviatilis (Diptera: Culicidae), and transcriptional changes associated with its native Wolbachia infection. BMC Genom 18(1):6–24. https://doi.org/10.1186/s12864-016-3441-4
Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW, Li RQ, Yang KY, Li J, Li M, Law PT, Wu YL, Cai ZL, Qin H, Bao Y, Leung RK, Ng PK, Zou J, Zhong XJ, Ran PX, Zhong NS, Liu ZG, Tsui SK (2015) The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J Allergy Clin Immunol 135(2):539–547. https://doi.org/10.1016/j.jaci.2014.09.031
Chen B, Zhang YJ, He Z, Li WS, Si FL, Tang Y, He QY, Qiao L, Yan ZT, Fu WB, Che YF (2014) De novo transcriptome sequencing and sequence analysis of the malaria vector Anopheles sinensis (Diptera: Culicidae). Parasite Vector 7(1):314–325. https://doi.org/10.1186/1756-3305-7-314
Chen W, Liu YX, Jiang GF (2015) De novo assembly and characterization of the testis transcriptome and development of EST-SSR markers in the cockroach Periplaneta Americana. Sci Rep 5:11144–11155. https://doi.org/10.1038/srep11144
Conte YL, Alaux C, Martin JF, Harbo JR, Harris JW, Dantec C, Séverac D, Cros-Arteil S, Navajas M (2011) Social immunity in honeybees (Apis mellifera): transcriptome analysis of varroa-hygienic behaviour. Insect Mol Biol 20(3):399–408. https://doi.org/10.1111/j.1365-2583.2011.01074.x
David JP, Faucon F, Chandor-Proust A, Poupardin R, Riaz MA, Bonin A, Navratil V, Reynaud S (2014) Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genom 15(1):174–188. https://doi.org/10.1186/1471-2164-15-174
Grbić M, Van Leeuwen T, Clark RM, Rombauts MS, Rouze R, Grbić V, Osborne EJ, Dermauw W, Ngoc PCT, Ortego F, Hernández-Crespo P, Diaz I, Martinez M, Navajas M, Sucena E, Magalhães S, Nagy L, MPace R, Djuranović S, Smagghe G, Iga M, Christiaens O, Veenstra JA, Ewer J, Villalobos RM, Hutter JL, Hudson SD, Velez M, Yi SV, Zeng J, Pires-daSilva A, Roch F, Cazaux M, Navarro M, Zhurov V, Acevedo G, Bjelica A, Fawcett JA, Bonnet E, Martens C, Baele G, Wissler L, Sanchez-Rodriguez A, Tirry L, Blais C, Demeestere K, Henz SR, Gregory TR, Mathieu J, Verdon L, Farinelli L, Schmutz J, Lindquist E, Feyereisen R, Van de Peer Y (2011) The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479(7374):487–492. https://doi.org/10.1038/nature10640
He ML, Xu J, He R, Shen NX, Gu XB, Peng XR, Yang GY (2016) Preliminary analysis of Psoroptes ovis transcriptome in different developmental stages. Parasite Vector 9(1):570–581. https://doi.org/10.1186/s13071-016-1856-z
Hoy MA, Yu FY, Meyer JM, Tarazona OA, Jeyaprakash A, Wu K (2013) Transcriptome sequencing and annotation of the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae): a cautionary tale about possible contamination by prey sequences. Exp Appl Acarol 59(3):283–296. https://doi.org/10.1007/s10493-012-9603-4
Hu L, Zhao YE, Yang YY, Niu DL, Wang RL, Cheng J, Yang F (2016) De novo RNA-seq and functional annotation of Sarcoptes scabiei canis. Parasitol Res 115(7):2661–2670. https://doi.org/10.1007/s00436-016-5013-6
Jiang X, Hall AB, Biedler JK, Tu Z (2017) Single molecule RNA sequencing uncovers trans-splicing and improves annotations in Anopheles stephensi. Insect Mol Biol 26(3):298–307. https://doi.org/10.1111/imb.12294
Kim K, Kim JH, Kim YH, Lee SH (2016) De novo transcriptome profiling and characterization of voltage-sensitive sodium channel gene of Tropilaelaps mercedesae parasitizing honey bees. J Asia Pac Entomol 19(1):89–93. https://doi.org/10.1016/j.aspen.2015.12.001
Li D, Liang Y, Wang X, Wang XW, Wang L, Qi M, Yu Y, Luan YY (2015) Transcriptomic analysis of Musca domestica to reveal key genes of the prophenoloxidase-activating system. G3 Genes Genom Genet 5(9):1827–1841. https://doi.org/10.1534/g3.115.016899
Lin C, Chen F, Yu SJ, Ding LL, Yang J, Luo R, Tian HX, Li HJ, Liu HQ, Ran C (2016) Transcriptome and difference analysis of fenpropathrin resistant predatory mite, Neoseiulus barkeri (Hughes). Int J Mol Sci 17(6):704–717. https://doi.org/10.3390/ijms17060704
Liu B, Jiang G, Zhang Y, Li JL, Li XJ, Yue JS, Chen F, Liu HQ, Li HQ, Li HJ, Zhu SP, Wang JJ, Ran C (2011) Analysis of transcriptome differences between resistant and susceptible strains of the citrus red mite Panonychus citri (Acari: Tetranychidae). PLoS ONE 6(12):1159–1160. https://doi.org/10.1371/journal.pone.0028516
Lv Y, Wang W, Hong S, Lei ZT, Fang FJ, Guo Q, Hu SL, Tian MM, Liu BQ, Zhang DH, Sun Y, Lei Ma, Shen B, Zhou D, Zhu CL (2016) Comparative transcriptome analyses of deltamethrin-susceptible and-resistant Culex pipiens pallens by RNA-seq. Mol Genet Genom 291(1):1–13. https://doi.org/10.1007/s00438-015-1109-4
Marco LD, EpisM S, Comandatore F, Porretta D, Cafarchia C, Mastrantonio V, Dantas-Torres F, Otranto D, Urbanelli S, Bandi C, Sassera D (2017) Transcriptome of larvae representing the Rhipicephalus sanguineus complex. Mol Cell Probe 31:85–90. https://doi.org/10.1016/j.mcp.2016.02.006
Meisel RP, Scott JG, Clark AG (2015) Transcriptome differences between alternative sex determining genotypes in the house fly, Musca domestica. Genome Biol Evol 7(7):2051–2061. https://doi.org/10.1093/gbe/evv128
Moreira HNS, Barcelos RM, Vidigal PMP, Klein RC, Montandon CE, Maciel TEF, Carrizo JFA, Lima PHC, Soares AC, Martins MM, Mafra C (2016) A deep insight into the whole transcriptome of midguts, ovaries and salivary glands of the Amblyomma sculptum tick. Parasitol Int 66(2):64–73. https://doi.org/10.1016/j.parint.2016.10.011
Olds BP, Coates BS, Steele LD, Sun W, Agunbiade TA, Yoon KS, Strycharz JP, Lee SH, Paige KN, Clark JM, Pittendrigh BR (2012) Comparison of the transcriptional profiles of head and body lice. Insect Mol Biol 21(2):257–268. https://doi.org/10.1111/j.1365-2583.2012.01132.x
Oleaga A, Obolo-Mvoulouga P, Manzanoromán R, Pérez-Sánchez R (2017) Functional annotation and analysis of the Ornithodoros moubata midgut genes differentially expressed after blood feeding. Ticks Tick Borne Dis 8(5):693–708. https://doi.org/10.1016/j.ttbdis.2017.05.002
Qu SX, Ma L, Li HP, Song JD, Hong XY (2016) Chemosensory proteins involved in host recognition in the stored-food mite Tyrophagus putrescentiae. Pest Manag Sci 72(8):1508–1516. https://doi.org/10.1002/ps.4178
Reid WR, Zhang L, Liu N (2015) Temporal gene expression profiles of pre blood-fed adult females immediately following eclosion in the southern house mosquito Culex Quinquefasciatus. Int J Biol Sci 11(11):1306–1313. https://doi.org/10.7150/ijbs.12829
Schicht S, Qi WH, Poveda L, Strube C (2014) Whole transcriptome analysis of the poultry red mite Dermanyssus gallinae (De Geer, 1778). Parasitology 141(3):336–346. https://doi.org/10.1017/S0031182013001467
Stuglik MT, Babik W, Prokop Z, Radwan J (2014) Alternative reproductive tactics and sex-biased gene expression: the study of the bulb mite transcriptome. Ecol Evol 4(5):623–632. https://doi.org/10.1002/ece3.965
Xu ZF, Zhu WY, Liu YC, Liu X, Chen QS, Peng M, Wang XZ, Shen GM, He L (2014) Analysis of insecticide resistance-related genes of the carmine spider mite Tetranychus cinnabarinus based on a De novo assembled transcriptome. PLoS ONE 9(5):e94779. https://doi.org/10.1371/journal.pone.0094779
Xu XL, Cheng TY, Yang H, Yan F, Yang Y (2015) De novo sequencing, assembly and analysis of salivary gland transcriptome of Haemaphysalis flava and identification of sialoprotein genes. Infect Genet Evol 32(1):135–142. https://doi.org/10.1016/j.meegid.2015.03.010
Zajac BK, Amendt J, Horres R, Verhoff MA, Zehner R (2015) De novo transcriptome analysis and highly sensitive digital gene expression profiling of Calliphora vicina (Diptera: Calliphoridae) pupae using MACE (massive analysis of cDNA ends). Forensic Sci Int Genet 15:137–146. https://doi.org/10.1016/j.fsigen.2014.11.013
Zhao YE (2017) A report of the humans bitten by Gamasid mites in the laboratory. Chin J Vector Biol Control 28(2):214 (in Chinese)
Zhou W, Russell CW, Johnson KL, Mortensen RD, Erickson DL (2012) Gene expression analysis of Xenopsylla cheopis (Siphonaptera: Pulicidae) suggests a role for reactive oxygen species in response to Yersinia pestis infection. J Med Entomol 49(2):364–375. https://doi.org/10.1603/ME11172
Zhou XJ, Qian K, Tong Y, Zhu JJ, Qiu XH, Zeng XP (2014) De novo transcriptome of the hemimetabolous German cockroach (Blattella germanica). PLoS ONE 9(9):e106932. https://doi.org/10.1371/journal.pone.0106932
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81471972 and 81271856).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Niu, D., Wang, R., Zhao, Y. et al. De novo RNA-seq and functional annotation of Ornithonyssus bacoti. Exp Appl Acarol 75, 191–208 (2018). https://doi.org/10.1007/s10493-018-0264-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-018-0264-9