Abstract
Extensive research has been conducted on the automatic classification of sleep stages utilizing deep neural networks and other neurophysiological markers. However, for sleep specialists to employ models as an assistive solution, it is necessary to comprehend how the models arrive at a particular outcome, necessitating the explainability of these models. This work proposes an explainable unified CNN-CRF approach (SleepXAI) for multi-class sleep stage classification designed explicitly for univariate time-series signals using modified gradient-weighted class activation mapping (Grad-CAM). The proposed approach significantly increases the overall accuracy of sleep stage classification while demonstrating the explainability of the multi-class labeling of univariate EEG signals, highlighting the parts of the signals emphasized most in predicting sleep stages. We extensively evaluated our approach to the sleep-EDF dataset, and it demonstrates the highest overall accuracy of 86.8% in identifying five sleep stage classes. More importantly, we achieved the highest accuracy when classifying the crucial sleep stage N1 with the lowest number of instances, outperforming the state-of-the-art machine learning approaches by 16.3%. These results motivate us to adopt the proposed approach in clinical practice as an aid to sleep experts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Sleep plays a vital role in an individual’s life and is crucial for mental and physical health. Conversely, sleep disorders or deficiencies can lead to chronic health problems, so it is essential to diagnose these problems early. Polysomnography (PSG) is the gold standard for sleep scoring and helps in the early diagnosis of many sleep-related disorders. PSG is mainly conducted in sleep clinics, and an individual usually sleeps there with multiple electrodes attached to the body, collecting multiple neurophysiological signals. The PSG signals are also prone to noise due to patients’ movements during the recordings. Next, the sleep experts label all the signals into different sleep stages by visually manipulating and understanding them; this process is known as sleep scoring. Until 2007, sleep experts used the Rechtschaffen and Kales (R and K rules) [21] manual for sleep scoring. Later, the manual was updated by the American Academy of Sleep Medicine (AASM) [2] for identifying different sleep stages.
Sleep stages are broadly categorized into two types, rapid eye movement (REM) and non-rapid eye movement (NREM) sleep stages. The NREM sleep stage is further classified into four stages: N1, N2, N3, and N4. NREM sleep constitutes around three-fourths of the total time spent in sleep, and REM sleep typically constitutes the remaining one-fourth of sleep. There is a cyclical transition between these two stages, and any irregularities in those cycles or an absence of a sleep stage may result in sleep disorders [15]; important features of EEG signals that are part of PSG can help in their identification. Some of the specific EEG characteristics of different sleep stages that a sleep expert looks for are shown in Fig. 1. Sleep stage N1 constitutes 2 to 5 percent of total sleep, with EEG containing rhythmic alpha waves, and is a transitional state in sleep architecture. Sleep stage N2 has sleep spindles and K-complexes in EEG and is between 40 and 60 percent of total sleep stages. Sleep stages N3 and N4 constitute 13 to 23 percent of total sleep, and EEG shows slow-wave activity. REM stages are characterized by sawtooth waves with theta and alpha activities on an EEG signal. In addition, muscle atonia and eye movements are present during this sleep stage [1].
All the sleep stages are interrelated, and the transition from one to another plays a significant role in the mental and physical development of human beings. Individuals who suffer from sleep disorders do not cycle through the regular stages of sleep. Thus, classifying each sleep stage with high accuracy, whether it is a short transition period of stage N1 or a more extended period of stage N2, plays a crucial role in identifying sleep disorders. Therefore, a sleep expert has to look into multiple EEG signals manually, and labeling the signals with sleep stages becomes labor-intensive and results in delayed and expensive results. Hence, to avoid this costly and intensive process, some researchers have been exploring methods for automating multi-class sleep stage classification using tools from artificial intelligence.
We rely on an intuitive idea of “explanation,” which refers to any hint that could assist the human decision-maker in comprehending the decision (in our work, the sleep stage identification). It is now vital for systems to provide an accurate decision and extra information that explains or supports the decision made by complex algorithms performing automated diagnosis tasks [9]. Unfortunately, most medical explainable artificial intelligence (XAI) research studies have focused on computer vision tasks and less often on time series. This work, which focuses on the latter type of data, aims to fill this gap in the literature. Another significant gap in previous research is that most studies have focused on increasing the overall accuracy of sleep stage identification and less on individual sleep stages.
In this paper, we propose an explainable deep learning approach for sleep stage identification (SleepXAI) that surpasses the existing methods in terms of accuracy and provides an explanation for the multi-class predictions by utilizing gradient-weighted class activation mapping (Grad-CAM) [22]. First, we focus on increasing the overall accuracy of the sleep stage identification task based on a single-channel EEG signal, specifically for sleep stage N1, where most of the previous work has demonstrated the lowest accuracy due to a low number of instances. In this regard, we utilized a combination of 1-D convolutional neural network (CNN) blocks (Sleep labeler) and conditional random fields (CRFs) [26] to classify the sleep labels by learning the temporal dependencies and contextual information and also calculated the probability scoring. Second, the proposed model exhibits the explainability of the obtained results using modified Grad-CAM, which generates a heatmap visualization of the univariate EEG signals. It highlights the specific characteristics of EEG signals, which explain the decision-making of the SleepXAI model. Hence, the proposed model can act as an automated assistive solution for sleep experts that reduces the manual labor involved and results in a timely prediction of sleep disorders.
We summarize the main contributions of our proposed approach as follows:
-
1.
We introduce a modified Grad-CAM for multi-class sleep labels to visualize a heatmap of specific features learned on a univariate EEG signal. The proposed method results in an explainable sleep stage classification, leading to human-readable output. The proposed approach can be used as an assistive solution in sleep laboratories, addressing the trust gap in the existing black-box models.
-
2.
The proposed approach demonstrates a higher overall classification accuracy than the comparative state-of-the-art deep learning algorithms. Moreover, it only requires a single-channel EEG signal instead of multiple signals for performing the classification while maintaining accuracy. Furthermore, the proposed approach achieved the highest accuracy when classifying the sleep stage N1, which has the lowest number of instances and serves as a transition stage from waking to sleeping.
The rest of the paper is organized as follows. Section 2 describes research related to the proposed method. The preliminaries and basic definitions are discussed in Section 3. The proposed explainable deep learning approach, SleepXAI, is explained in Section 4. The performance analysis of the proposed study is presented in Section 4. The experimental results are demonstrated and discussed in Section 5. Finally, Section 6 concludes the work along with a discussion of the future aspects of the proposed study.
2 Literature review
In this section, we review the research literature on the following two categories:
-
1.
Sleep stage classification;
-
2.
Explainable machine learning in healthcare.
2.1 Sleep stage classification
Several studies have recently been conducted to automate the task of sleep stage classification based on machine learning. These studies can be divided into traditional and machine-learning approaches. In the traditional approach, raw PSG signals are extensively preprocessed to transform them for feature extraction. After that, the various features related to the time domain, frequency domain, and linear features are extracted with the help of prior knowledge from sleep experts. These extracted features are then fed into traditional machine learning classifiers for the sleep stage classification task. For example, [7] extracted the time-frequency representation (TFR) images using the Fourier-Bessel decomposition method (FBDM) and a CNN classifier to classify a publically available library of sleep EEG signals. Using EEG signals, the created classification system has obtained a classification accuracy of 91.90% for classifying six distinct sleep stages. Similarly, [29] extracted the time, frequency, and fractional Fourier transform (FRFT) domain features from a single-channel EEG and fed those features into bidirectional Long short-term memory (LSTM) to learn the rules for transitioning between sleep stages. The proposed approach resulted in an overall accuracy of 81.6% for the Fpz-Cz EEG channel of Sleep-EDF. Many studies have thus been conducted based on this approach, and the accuracy is relatively high. However, there are many drawbacks related to these approaches. The main drawbacks are the requirement for extensive preprocessing and the domain expertise for feature engineering. Moreover, this approach lacks a generalized solution for automated sleep stage classification due to the diversified sleep patterns of different patients.
The second approach is based on machine learning, where deep learning models are implemented to extract the features and classify them into respective sleep labels. This approach can be further classified based on multiple or single-channel PSG signals. For example, [24] explored CNN for sleep stage classification based on EEG and EOG signals and obtained an accuracy of 81.0%. Similarly, [10] used a DeConvolutional Neural Network (DCNN) that inversely maps features of a hidden layer back to the input space to predict the sleep stage label at each timestamp using a multivariate time series of PSG recordings. These recordings included six channels and two leg electromyogram channels. Much research has been conducted based on multiple-channel PSG signals, and state-of-the-art accuracy has been achieved, but some drawbacks deserve mention. The main drawback is that subjects have to sleep with different electrodes and wires attached to their bodies, which affects sleep and introduces noise.
Nevertheless, multiple-channel-based approaches are effective in the sleep laboratory environment, while the single-channel approach is highly productive when designing home-based sleep monitoring systems. Dut et al. [3] used a time-distributed convolutional network architecture to extract the features, capture the temporal information, and label sleep stages with an accuracy of 85.0% from a single raw-channel EEG signal. Eldele et al. [4] used the multi-resolution convolutional neural network (MRCNN) and adaptive feature recalibration (AFR) to extract the features from the EEG channel. Then, they fed those into the temporal context encoder (TCE) module, which captures the temporal dependencies among the extracted features using the multi-head attention (MHA) mechanism, achieving an overall accuracy of 85.6% on the Sleep-EDF dataset. Yang et al. [28] introduced 1D-CNN-HMM, an automatic sleep stage categorization model based on a single EEG channel. A deep one-dimensional convolutional neural network (1D-CNN) and a hidden Markov model are combined in the 1D-CNN-HMM model (HMM). Experimental results showed that the overall accuracy and kappa coefficient of 1D-CNN-HMM on Fpz-Oz channel EEG from the Sleep-EDFx dataset could reach 83.98% and 78.0%, respectively. [16] developed EEGSNet, a deep learning model based on CNNs and two-layer bidirectional Long short-term memory networks (Bi-LSTM) to learn the transition rules and characteristics from neighboring epochs and classify sleep stages with an accuracy of 86.82%.
2.2 Explainable machine learning in healthcare
The literature review has shown that most research for interpretation and explanation has been conducted on 2D- CNN models in the medical domain, leaving a gap in explanation in the medical time series data. Jiang et al. [11] proposed a Grad-CAM-based multi-label classification model that automatically locates different retinopathy lesions using original DR fundus images. Li et al. [17] introduced a novel recurrent-convolution network for EEG-based intention recognition. Grad-CAM was used for the channel selection to omit the unnecessary information produced by redundant channels. Hata et al. [8] proposed the classification of aortic stenosis using ECG images and explored the relationship between the trained network and its determination using Grad-CAM.
The primary motivation for conducting this study was to overcome the mentioned drawback. First and foremost, we created an architecture that can label sleep stages based on a single-channel EEG signal, achieving state-of-the-art accuracy with a primary focus on sleep stage N1, which in most literature reviews has the lowest accuracy. Second, we designed an architecture that can explain the model outcome from univariate time-series data for multiple classes using Grad-CAM, making it easier for patients and doctors to interpret the results. Furthermore, we proposed structural changes to Grad-CAM to interpret the univariate data, improve EEG data visualization, and compare the results with existing models.
3 Preliminaries
This section presents the basic definitions which were utilized to develop the CNN-CRF model and which further support the establishment of the SleepXAI architecture. Furthermore, the motivation behind considering the CRF is discussed in this section.
3.1 Basic definitions
The naive Bayes classifier, which is a generative algorithm, indicates that to predict a class label y using a naive Bayes algorithm, based on the independence assumption \(P\left (s_{t}\mid y, s_{1}...s_{t-1},s_{t+1} ...s_{t} \right ) = P\left (s_{t}\mid y \right )\), we can decompose the conditional probability to solve
where
However, in a discriminative model, such as the logistic regression classifier, we replace P(y∣s) with the Bayes equation:
Since the denominator P(s) does not contribute any information in the argmax term, we obtain
which is equivalent to the product of the prior and the likelihood. It must be observed that P(s∣y)P(y) = P(s,y), the joint distribution of s and y. Thus, by learning the conditional probability distribution in discriminative models, the decision boundary can be learned for classification. Therefore, given an input point, it can use the conditional probability distribution to identify its class.
3.2 Motivation for conditional random fields
As discussed above, we can model the conditional distribution as follows:
In CRFs, input data are processed sequentially. Furthermore, to model this behavior, we will use feature functions that combine multiple input vectors s and additional information, namely:
-
1.
The position t of the data point to be predicted;
-
2.
The label yt− 1 of data point t − 1 in input vector s;
-
3.
The label yt of data point t in s.
Thus, the feature function can be written as Φ(s,yt− 1,yt). The purpose of the feature function is to express the characteristics of the input sequence. To build the conditional field, we next assign each feature function a set of weights (lambda values) which the algorithm is to learn.
where the partition function Z(s) is
In summary, we use CRFs by first defining the feature function needed, initializing the weights to random values, and then applying gradient descent iteratively until the parameter values (in this case, lambda) converge. We can see that CRFs are similar to logistic regression since they use the conditional probability distribution, but we extend the algorithm by applying feature functions as our sequential inputs. The process of integrating the λ and learning the lambda using iterative gradient descent for the proposed CNN-CRF model is further explained in Section 4.
4 Proposed SleepXAI architecture
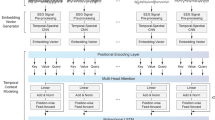
An illustration of the proposed SleepXAI architecture is presented in Fig. 2. It consists of three main parts: 1) sleep encoder, 2) sleep labeler, and 3) N-class modified Grad-CAM. First, the 30-second (time period) raw EEG signal is fed into the sleep encoder part for feature extraction using the time distributed layer. Next, the extracted features are fed into the sleep labeler part, which utilizes the benefits of both CNN and CRF layers to learn the temporal dependencies with contextual information and generates the probability scoring among different sleep labels. The sleep encoder and sleep labeler network parameters employed in SleepXAI can be seen in Table 1. Finally, we implement the Grad-CAM part, which exploits the spatial information preserved through the convolution layers, to create the heat visualization of the characteristics of EEG signals that are vital for a classification decision. The details of the primary motivation behind the proposed architectural design of the sleep encoder, sleep labeler, and Grad-CAM are discussed in the next section.
4.1 Sleep encoder
The sleep encoder consists of convolution, maxpool, spatial dropout, and global maxpool layers. The convolution layer is implemented in pairs to extract specific characteristics from the input EEG signal. The first layer extracts low-level features, and the second allows the network to extract high-level features. 1D-CNN performs convolution operations on EEG signals to obtain one-dimensional features, and various kernels extract unique EEG characteristics. The forward propagation in 1D-CNN is expressed as follows [14]:
\({b_{k}^{l}}\) is kernel bias, \(s_{i}^{l-1}\) is the output of the ith neuron at layer l − 1, \(w_{ik}^{l-1}\) is the kernel from the ith neuron at layer l − 1 and the kth neuron at layer l
where \({y_{k}^{l}}\) is defined as the intermediate output, and the activation function is denoted by f(⋅).
After that, the Max pooling operation is applied to downsample the feature maps generated by the filter by selecting the maximum value, thus retaining the most prominent features of the previous feature map and reducing the dimensionality. Spatial Dropout [27] enhances independence between feature maps by eliminating complete 1D feature maps in favor of single parts when feature maps are highly correlated. The global maxpool block is embedded after the last convolutional layer to downsample the input by taking the maximum value over the time dimension. The dropout operation enhances the generalization of the neural network model by randomly selecting the neurons from the model, solving the overfitting problem. Finally, we have implemented a dense layer in the SleepXAI network, which receives output from every neuron in its preceding layer, changing the encoded sequence dimensions. The primary function of the sleep encoder is to convert the time-invariant patterns from the raw input time series EEG signals into an encoded sequence, making it easier for the sleep labeler to learn long temporal dependencies.
4.2 Sleep labeler
Sleep cycles throughout the night exhibit specific transitions from one sleep stage to another [15], and the sleep labelers attempt to capture these time-related transitions by learning them. The encoded sequence from the sleep encoder is fed into the sleep labeler, which identifies the temporal information, such as transition rules, and generates the probability scoring of each sleep label utilizing the implementation of both CNN and CRF in a unified manner. A detailed description of the CNN-CRF model and its integration with the proposed problem scenario is explained first in this section. Next, the development of explainable sleep stage identification using Grad-CAM is described in this section. Furthermore, an algorithm is presented which enumerates the whole process of the proposed SleepXAI approach.
4.2.1 CNN-CRF model integration with SleepXAI
As described in Section 3, generative architectures model the joint probability distribution p(s,y) for classification, where y = [y1,y2,...,yT] is the output label vector and s = [s1,s2,...,sT] are input feature vectors. In the proposed approach, the output labels consist of five sleep stages, namely, \(y_{t} \in \left \{ W, N_{1}, N_{2}, N_{3}, REM \right \}\).
A general classification model without considering the dependencies between the adjacent feature vectors and different sleep stages is shown in Fig. 3. With this consideration of independence between adjacent feature vectors st− 1 and st+ 1, the predictive model assigns a probability to each label yt given its associated input St, and the conditional probability distribution for that is given by
However, as discussed in the Introduction, there are dependencies between different sleep stages that can be modeled and identified by utilizing the CRF properties and by learning the inter-dependencies of the input sequence of 30-second time periods and feature vectors in linear chains. An illustration of this is presented in Fig. 4.
A mathematical representation of Fig. 4 can be given as follows
To implement this CRF model using neural networks, a set of weights λ can be assigned to feature vectors as described earlier in (6), which can be learned or estimated by neural networks. To estimate the parameters (lambda), maximum likelihood estimation is utilized and applied to the negative log of the distribution, in order to make the partial derivative easier to calculate, which is given by
To apply maximum likelihood to the negative log function, we take the argmin (because minimizing the negative will yield the maximum). To find the minimum, we can obtain the partial derivative with respect to lambda, and get:
We use the partial derivatives as a step in gradient descent; furthermore, in each incremental step of λ, an update is given by
This process of gradient descent occurs iteratively to update parameter λ, with a small step, until the values converge.
Thus, a CRF layer is added at the end of the sleep labeler in the SleepXAI architecture. CRFs act as a probabilistic graph model used to predict the sequences that use the contextual information from a neighboring sample to add information that the model uses to make correct predictions. Therefore, in the context of sleep stage classification, certain sleep stages are followed or perceived by certain sleep stages. These transitions are very well learned by CRFs, making the accuracy of the classification of classes with small instances such as N1 and N3 much higher.
4.3 Explainable sleep stage classification using N-class modified Grad-CAM
We utilized the well-known explainable model named Grad-CAM, which utilizes class activation maps (CAMs) to recognize the heatmaps in the input data. The CAM for a particular class c1 is the weighted-sum of activation maps (A1,A2,...,Ak) generated from k convolution filters, which is given by
An illustration of CAMs is shown in Fig. 5. Furthermore, global average pooling is applied over each activation map, which is given by
Thus, a class label is identified based on the following equation:
A general vector-form for CAM is given as follows:
where \(A_{ij}^{k}\) is the pixel at the (i,j) location of the k th feature map.
From Grad-CAM, it is established that gradients of the last layer in the NN model led to weights \({w_{c}^{k}}\) of the classes. Thus, we can be computationally efficient while training the model. This established fact is also described herein.
From CAM, we obtained Yc, Furthermore, let \(F^{k} = \frac {1}{Z}{\sum }_{i}^{}{\sum }_{j}^{}A_{ij}^{k}\); then, \(Y^{c} = {\sum }_{k}^{}{w_{k}^{c}}\cdot F^{k}\); we then have:
which turns out to be
Furthermore, we can write
By rearranging the above equation, we obtain
Using gradient flowing from the output class into the activation maps of the last convolutional layer as weight (\({w_{c}^{k}}\)) w.r.t class c, we do not need to perform retraining to obtain weights, which is a potential reason to integrate Grad-CAM with the proposed SleepXAI architecture.
For integrating Grad-CAM in the proposed model, the multi-class labeled hypnograms obtained from the sleep labeler are fed into Grad-CAM. To realize Grad-CAM, the input is fed to a set of convolutional layers and class activation maps. This provides us with coarse localization maps highlighting the crucial waves corresponding to particular sleep stages, which makes it a self-explanatory model named SleepXAI herein.
5 Experimental results
In this section, we first discuss the dataset used for the performance evaluation of the proposed approach. Furthermore, different performance metrics are described. Then, we demonstrate the results and discuss the accuracy and explainability achieved by the proposed SleepXAI architecture. Furthermore, several existing methods are considered for comparative analysis.
5.1 Dataset
In this study, we evaluated our model on the Sleep-EDF (Sleep-EDFx, 2013 version) [6, 13] dataset and compared its performance with other state-of-the-art algorithms in Section 5.4. The dataset consists of two subsets: (1) the Sleep Cassette (SC) subset of 20 healthy participants aged 25–34 to explore the effects of age on sleep; and (2) the Sleep Telemetry (ST) subject of 22 Caucasian people to study temazepam’s effects on sleep. We used the two EEG signals (Fpz-Cz and Pz-Oz) extracted from the PSG of the SC subset, which contains two nights of sleep recorded for each subject, except for subject 13, who had only the first night recorded. Sleep experts manually annotated the recordings into specific sleep labels. There were long periods of wakefulness during the beginning and end of each recording, and periods of body movement and unclassified periods were omitted from the data used in the evaluation process. Each signal was annotated with its specific sleep label based on a 30-second window called an epoch, and the total number of epochs is shown in Table 2.
5.2 Evaluation approach and metrics
There are two primary evaluation approaches to assess a machine learning approach deployed in the medical domain. The first one is the intra-patient approach, where the data from the same subject can be used during the training and testing of the model. In the second inter-patient approach, the data for training and testing the model comes from different subjects. Since the dataset on which the model is trained in the medical domain differs from the dataset on which it is evaluated, the inter-patient approach is a more practical assessment approach. Therefore, we used the inter-patient approach in this study to assess the proposed approach’s performance.
To implement this approach, we used k-fold cross-validation, with k set to 20 based on the total number of subjects. Each subject has two nights of sleep recording, except for one subject with a single night of sleep recording. Each recording contains a whole night of EEG data split into 30-second time windows called epochs. We used the sleep recordings of 19 subjects to train and validate the model, and the remaining subjects’ recordings to test the trained model. This procedure was repeated 20 times so that the model could be evaluated against each subject. Then, we aggregated the results from each fold and computed the performance using the evaluation metrics described below.
We used overall evaluation metrics such as accuracy, precision, recall (sensitivity), and the F1 score (per class) to assess the proposed approach performance.
where TP = true positive (predicted output true and actual output also true), FP = false positive (predicted output true and actual output false), TN = true negative (predicted output false and actual output also false), and FN= false negative (predicted output false and actual output true).
Another performance evaluation metric used was the F1 score, which is given by
where
and
5.3 Performance analysis
The performance of the SleepXAI approach was evaluated on the two EEG signals (Fpz-Oz and Pz-Oz) of the PSG recordings of the Sleep-EDF dataset. In addition, we also compared the sleep stage classification of accuracy between CNN-CNN and CNN-CRF implementation in the sleep labeler. Tables 3, 4, 5, and 6 demonstrate the classification report of both these architectures on the respective signals.
The CNN-CRF model has shown a higher accuracy for classifying different sleep stages for both the signals when compared with the CNN-CNN model. For example, the SleepXAI achieved the highest accuracy of 86.81% on the EEG Fpz-Cz channel, which is around 2% more than the CNN-CNN implementation. This may be because there are specific transitions that the sleep stages follow during the complete sleep cycle. The CRF layer can learn this contextual information by considering the neighboring sleep stages, thus increasing the amount of information to make current sleep label predictions. Furthermore, it helps the CNN-CRF model capture the sleep stages with fewer instances with higher accuracy. For example, an accuracy of 63% was achieved by SleepXAI for sleep stage N1, which is 15% more than that achieved with the CNN-CNN implementation.
The CNN-CNN and CNN-CRF implementations showed higher classification accuracy on the EEG Fpz-Cz signal than on the EEG Pz-Oz signal. We can hypothesize that this can be due to electrode placement for both these channels [12]. Certain characteristics are present in specific sleep stages, such as K-complexes, delta activities, and sleep spindles, which predominantly occur in the brain’s frontal lobe, and the Fpz-Cz channel placed in the frontal region can easily capture them. Furthermore, alpha activities are occipital phenomena, but they can easily manifest themselves in the brain’s frontal area, allowing the Fpz-Cz channel to capture them. On the other hand, the placement of the Pz-Oz electrode is on top of the parietal region of the brain, and theta and higher-frequency sleep spindle activity are mostly parietal phenomena. Therefore, the presence of theta activities in the multiple sleep stages does not make it beneficial when distinguishing between different sleep classes lowering the classification accuracy.
Figure 6 shows the confusion matrices of CNN-CNN and CNN-CRF implementation on the Fpz-Cz and Pz-Oz channels, and SleepXAI demonstrates the highest accuracy on the Fpz-Cz channel, as shown in Fig. 6(b). We will discuss the potential reason for most misclassified sleep stage pairs according to the AASM sleep scoring transition rules [2]. Both the sleep stages N1 and Wake have shown misclassification due to similarity in EEG characteristics, alpha, and low-voltage activities. In addition, the sleep spindles are among the defining characteristics of sleep stage N2, but these spindles can persist in sleep stage N3, contributing to misclassification between these two stages. The EEG characteristic K-complexes can be seen in both sleep stages, N1 and N2. The classification between these two classes mostly depends on the body and slow eye movements, which are difficult to capture with EEG signals, leading to misclassification between the pair. The sleep stages N1 and REM demonstrate mixed EEG frequency rates and low amplitude, making the classification difficult.
Figure 7 shows the comparison between the hypnogram of the sleep cycle of two nights of sleep of two different subjects annotated by the sleep expert and predicted by the CNN-CRF (SleepXAI approach) and CNN-CNN. It can be clearly seen that the CNN-CRF model captures the sleep stages with a lower number of instances due to minimal transition periods. As a result, there was a significant increase in the prediction of sleep stage N1, which can be seen in Table 7. The F1 score for detecting sleep stage N1 by CNN-CRF was around 95.34%, more than 30.82% greater than that by CNN-CNN. Similar results can be seen in the classification of other sleep stages. This demonstrates that the unified CNN-CRF approach increased the accuracy.
5.4 Comparison with other state-of-the-art algorithms
To further evaluate the performance of the SleepXAI approach, we compared the results achieved by other state-of-the-art sleep stage classification methods. Table 8 compares our proposed approach and these methods across total accuracy and F1 score for each class. In addition, we compared the performance with two different channels, EEG Fpz-Cz and Pz-Oz. Again, the results showed that our proposed approach achieved the highest overall accuracy and significantly increased the accuracy of detecting sleep stage N1 for both channels.
5.5 Analysis of Grad-CAM visualizations
The explainable part of the SleepXAI approach is the implementation of the Grad-CAM. This section demonstrates the result obtained by Grad-CAM for multi-label sleep stage classification. Using Grad-CAM, we can visually validate where the SleepXAI model is looking in the EEG signal and verify that it looks at the correct EEG characteristics mentioned in the manual scoring of the sleep stages in the AASM manual. In addition, Table 9 shows specific characteristics of the 30-second time period that a sleep expert looks into for scoring specific sleep labels. Finally, Fig. 8 demonstrates the output of the Grad-CAM in the SleepXAI approach, highlighting the regions of the signal which are impacted most when making the classification decision using the proposed approach.
The sleep stage is scored as Wake by a sleep expert when more than 50% of the time period has an alpha rhythm, and it can be clearly seen that the Grad-CAM output when SleepXAI distinguishes a time period as Wake has most of the alpha rhythm as highlighted regions. For a time period to be classified as sleep stage N1, there are theta waves (4-7Hz) and vertex sharp waves. The same regions are highlighted by Grad-CAM when making a classification decision of sleep stage N1. The sleep stage N2 has the presence of K complexes and one or more sleep spindles, and the SleepXAI model also emphasizes these regions when classifying a time period as sleep stage N2, which is validated by Grad-CAM output. The sleep stage N3 time period, also known as deep sleep, has 20% or more regions containing slow-wave activity. The same regions are highlighted by the Grad-CAM output of the SleepXAI model when it classifies a time period as sleep stage N3. Finally, the REM stage has the presence of sawtooth waves, and the sawtooth waves are captured by the proposed approach when making the classification decision of REM.
For the ML model to be implemented in the clinical environment, some explainability parts must emphasize why the decision has been made. For example, from the output generated by the Grad-CAM part of the SleepXAI approach, a sleep expert can validate whether the model looks into the same EEG characteristics they look for while scoring the sleep stages.
6 Conclusion and future work
Artificial intelligence can be crucial in automating and revolutionizing the health sector. In this context, this study proposed an explainable deep learning approach named SleepXAI, which performs the automatic classification of multiple sleep stages. To the best of our knowledge, this is the first study to report the characteristics influencing the classification decisions of deep models for the multi-class classification of sleep stages only using single-channel EEG signals. The SleepXAI introduces explainability by generating a heatmap visualization of salient features learned for the predicted sleep stage on a univariate EEG signal. It allows the sleep experts to correlate the learned features visually with the AASM manual sleep scoring rules, thus improving trust in black box systems with explanations. This study substantially contributes to the medical field by explaining decision-making and can act as an aid to clinicians.
As shown in Table 8, the proposed approach outperforms other state-of-the-art sleep-scoring algorithms based on the Sleep-EDFx 2013 dataset. However, this dataset is too small, and a much larger dataset is necessary to test the model’s performance in terms of explainability and accuracy. In addition, a diversified dataset comprising data collected over a more extended period to simulate emotional, stress, and health circumstances is required to demonstrate the algorithm’s adaptability. Furthermore, the dataset should be compiled from a large geographical area to include individuals of various races and ethnicities. Finally, the algorithm has been tested on only two EEG signals (Fpz-Cz and Pz-oz) and must be tested on the other physiological signals collected during PSG. The adaptability and robustness of the proposed approach are currently being explored against other publically available datasets based on different physiological signals collected during PSG. The model interpretability and other practical challenges can be explored as an extension of the proposed approach in future work.
Data Availability
The data supporting this study’s findings are openly available in Sleep-EDF Database at https://www.physionet.org/content/sleep-edfx/1.0.0/RECORDS-v1 Sleep-EDF
Code Availability
The code supporting this study is available at https://github.com/michealdutt/SleepXAI SleepXAI
References
Acharya UR, Bhat S, Faust O et al (2015) Nonlinear dynamics measures for automated eeg-based sleep stage detection. Eur Neurol 74(5-6):268–287
Berry RB, Brooks R, Gamaldo CE et al (2012) The aasm manual for the scoring of sleep and associated events. rules, Terminology and Technical Specifications, Darien, Illinois. American Academy of Sleep Medicine 176:2012
Dut M, Goodwin M, Omlin CW (2021) Automatic sleep stage identification with time distributed convolutional neural network. In: 2021 International joint conference on neural networks (IJCNN), IEEE, pp 1–7
Eldele E, Chen Z, Liu C et al (2021) An attention-based deep learning approach for sleep stage classification with single-channel eeg. IEEE Trans Neural Syst Rehabilitation Eng 29:809–818
Ghimatgar H, Kazemi K, Helfroush MS et al (2019) An automatic single-channel eeg-based sleep stage scoring method based on hidden markov model. J Neurosci Methods 324:108,320
Goldberger AL, Amaral LA, Glass L et al (2000) Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation 101(23):e215–e220
Gupta V, Pachori RB (2021) Fbdm based time-frequency representation for sleep stages classification using eeg signals. Biomed Signal Process Control 64:102,265
Hata E, Seo C, Nakayama M et al (2020) Classification of aortic stenosis using ecg by deep learning and its analysis using grad-cam. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC), IEEE, pp 1548–1551
Holzinger A, Langs G, Denk H et al (2019) Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip Rev: Data Min Knowl Discov 9(4):e1312
Huang X, Shirahama K, Li F et al (2020) Sleep stage classification for child patients using deconvolutional neural network. Artif Intell Med 110:101,981
Jiang H, Xu J, Shi R et al (2020) A multi-label deep learning model with interpretable grad-cam for diabetic retinopathy classification. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC), IEEE, pp 1560–1563
Jobert M, Poiseau E, Jähnig P et al (1992) Topographical analysis of sleep spindle activity. Neuropsychobiology 26(4):210–217
Kemp B, Zwinderman AH, Tuk B et al (2000) Analysis of a sleep-dependent neuronal feedback loop: the slow-wave microcontinuity of the eeg. IEEE Trans Biomed Eng 47(9):1185–1194
Kiranyaz S, Avci O, Abdeljaber O et al (2021) 1d convolutional neural networks and applications: A survey. Mech Syst Signal Process 151:107,398
Kishi A, Struzik ZR, Natelson BH et al (2008) Dynamics of sleep stage transitions in healthy humans and patients with chronic fatigue syndrome. Am J Physiol Regul Integr Comp Physiol 294(6):R1980–R1987
Li C, Qi Y, Ding X et al (2022) A deep learning method approach for sleep stage classification with eeg spectrogram. Int J Environ Res Public Health 19(10):6322
Li Y, Yang H, Li J et al (2020) Eeg-based intention recognition with deep recurrent-convolution neural network: Performance and channel selection by grad-cam. Neurocomputing 415:225–233
Phan H, Andreotti F, Cooray N et al (2018) Dnn filter bank improves 1-max pooling cnn for single-channel eeg automatic sleep stage classification. In: 2018 40th annual international conference of the IEEE engineering in medicine and biology society (EMBC), IEEE, pp 453–456
Phan H, Andreotti F, Cooray N et al (2018) Joint classification and prediction cnn framework for automatic sleep stage classification. IEEE Trans Biomed Eng 66(5):1285–1296
Qu W, Wang Z, Hong H et al (2020) A residual based attention model for eeg based sleep staging. IEEE J Biomed Health Inform 24(10):2833–2843
Rechtschaffen A (1968) A manual for standardized terminology, techniques and scoring system for sleep stages in human subjects. Brain information service
Selvaraju RR, Cogswell M, Das A et al (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626
Seo H, Back S, Lee S et al (2020) Intra-and inter-epoch temporal context network (iitnet) using sub-epoch features for automatic sleep scoring on raw single-channel eeg. Biomed Signal Process Control 61:102,037
Sokolovsky M, Guerrero F, Paisarnsrisomsuk S et al (2019) Deep learning for automated feature discovery and classification of sleep stages. IEEE/ACM Trans Comput Biol Bioinform 17(6):1835–1845
Supratak A, Dong H, Wu C et al (2017) Deepsleepnet: a model for automatic sleep stage scoring based on raw single-channel eeg. IEEE Trans Neural Syst Rehabilitation Eng 25(11):1998–2008
Sutton C, McCallum A et al (2012) An introduction to conditional random fields. Found. Trends® Mach. Learn 4(4):267–373
Tompson J, Goroshin R, Jain A et al (2015) Efficient object localization using convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 648–656
Yang B, Zhu X, Liu Y et al (2021) A single-channel eeg based automatic sleep stage classification method leveraging deep one-dimensional convolutional neural network and hidden markov model. Biomed Signal Process Control 68:102,581
You Y, Zhong X, Liu G et al (2022) Automatic sleep stage classification: a light and efficient deep neural network model based on time, frequency and fractional fourier transform domain features. Artif Intell Med 127:102,279
Funding
Open access funding provided by University of Agder
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dutt, M., Redhu, S., Goodwin, M. et al. SleepXAI: An explainable deep learning approach for multi-class sleep stage identification. Appl Intell 53, 16830–16843 (2023). https://doi.org/10.1007/s10489-022-04357-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-04357-8