Abstract
The accuracy of graph based learning techniques relies on the underlying topological structure and affinity between data points, which are assumed to lie on a smooth Riemannian manifold. However, the assumption of local linearity in a neighborhood does not always hold true. Hence, the Euclidean distance based affinity that determines the graph edges may fail to represent the true connectivity strength between data points. Moreover, the affinity between data points is influenced by the distribution of the data around them and must be considered in the affinity measure. In this paper, we propose two techniques, CCGAL and CCGAN that use cross-covariance based graph affinity (CCGA) to represent the relation between data points in a local region. CCGAL also explores the additional connectivity between data points which share a common local neighborhood. CCGAN considers the influence of respective neighborhoods of the two immediately connected data points, which further enhance the affinity measure. Experimental results of manifold learning on synthetic datasets show that CCGA is able to represent the affinity measure between data points more accurately. This results in better low dimensional representation. Manifold regularization experiments on standard image dataset further indicate that the proposed CCGA based affinity is able to accurately identify and include the influence of the data points and its common neighborhood that increase the classification accuracy. The proposed method outperforms the existing state-of-the-art manifold regularization methods by a significant margin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Manifold learning and regularization methods have been widely used for data representation and processing respectively. A given data is represented as a weighted graph where the weights represent the similarity between data points. In an ideal case, similar data points should have a higher affinity, which satisfies the manifold assumption. In manifold learning, clusters of data points having larger affinity are kept spatially close when projected to lower dimensional space. Similarly, in manifold regularization, the labels of data points in a large affinity neighborhood are assumed to be similar and, hence, the approximated function is penalized of it changes label in such a neighborhood. This shows that affinity fundamental to both manifold learning and regularization methods, which also influence the accuracy of the underlying model.
Any manifold learning or regularization technique can be decomposed into three steps. (1) computation of the pairwise affinity, (2) creation of the graph, and (3) project the data into low dimensional space or approximate the generative function and penalize. When only data points are given, then an appropriate metric needs to be evolved to capture the similarity between data points. This is crucial as the efficiency of a learning technique hinges on the affinity metric. The affinity metric must be able to capture the dependence between data points and account for uneven sampling, if any, of the data points. Once the affinity is determined, geodesic using the heat kernel may be computed between data points assumed to be lying on the Riemannian manifold. The heat kernel enforces the smoothness assumption, which depends on the kernel bandwidth (σ).
The heat kernel affinity is governed by its bandwidth or scaling, which drastically reduces the affinity value for data points assumed to be distant or non-similar. The local scaling method further optimizes this bandwidth to fit the local neighborhoods of connected data points [1]. It often takes the distance between the point of interest and its 7th neighbor as affinity bandwidth, which suggests an increase in accuracy as compared to baseline methods. Instead of finding affinity from the given data points, [2] proposes to recreate optimal affinity assuming the graph partition has been provided. The removal of noisy edges that artificially increased or decreased the affinity between data points based on the maximum cliques further enhanced the manifold’s modeling[3]. A slightly different approach introduced in the paper [4] that combined the similar information from various neighborhoods to enhance the robustness of k NN. An adaptive neighborhood assigning technique [5] proposed to learn the graph adjacency. It can extensively explore the local manifold structure but fails to decode the global structure of the manifold. A different approach introduced by [6] that used the information theoretic approach to decode the similarity information. These methods directly rely on the Euclidean distance to construct a data graph that assumes the knowledge of the extent of the locally linear region and remains susceptible to noisy and highly correlated features.
In this work, we propose cross-covariance based graph affinity (CCGA) for the affinity approximation task. CCGA aims to preserve both linear and non-linear properties of the manifold accurately. The method initiates by mapping the data points into reproducing kernel Hilbert space (RKHS) using an appropriate kernel on the vertices of the underlying graph, which is followed by similarity measure between data points identified as neighbors in ambient space using Hilbert-Schmidt independence criterion (HSIC). The cross-covariance based affinity metric minimizes the effect of noise and error measurement by making it independent of its existing spatial locations. CCGA for the local region (CCGAL) method aims to strengthen the affinity by discovering the connectivity between data points, which may or may not be immediately connected but share one or more common local neighborhoods that indicate an innate similarity. Additionally, CCGA for the neighborhood (CCGAN) weighs the influence of respective neighbors of two connected data points for a correct measure of similarity.
The major contributions of the paper can be stated as, we propose Cross-Covariance based Graph Affinity between two data points that incorporates nonlinear dependence, the influence of neighboring data points, and the neighborhoods around data points. The first technique CCGAL, try to include the influence of the data points in the common neighborhood in the affinity metric between two points. The second technique CCGAN, takes into consideration the influence of the neighborhoods of data points that are adjacent to both the points. Table 1 describes the important mathematical symbols used in this paper.
The rest of the paper is organized as follows: Section 2 describes the existing literature that highlights state-of-the-art techniques and various affinities used in machine learning techniques. Section 3 illustrates the proposed technique and describes how measuring the dependence between the data points leads to a more accurate affinity. Sections 3.2 and 3.3 explains the affinity obtained based similarity estimation between two data in a local region and between two neighborhoods, respectively. Section 4 highlights the experiments performed on various real world and synthetic data to evaluate the performance of our techniques in terms of classification accuracy and dimensionality reduction. Section 5 concludes the findings and robustness of our techniques.
2 Related work
It is known that the affinity between data points depends not only on the location but also on the neighborhoods of two points. The density of data has been considered by various metrics in supervised [7], unsupervised [8], semi-supervised [9] and deep learning [10]. The affinity between two data points with different neighborhood density is different from the affinity when both neighborhoods have the same density [11]. Many of the existing literature shows that they adjust local data structures to improve the characteristics of the affinity graph. However, these techniques are highly susceptible to the occurrence of noise and irrelevant features. They mainly focus on the parameter σ of the RBF heat kernel to determine the similarity between two data points. To solve such issues, the graph-theoretic approach is applied by [3] that consider rebuilding the tight neighborhood by picking maximal clique, which in other words, maximizes the average pairwise similarity. The use of contextual information is proposed in [12] to obtain affinity scores between the pairs of data points. The similarity information propagates as diffusion on the graph. The SSNMM-isomap framework [13] employs LLE to preserve the optimal local properties that minimizes the pairwise distances between intraclass data points that assumed to lie in the same manifold. The inter-class distances between data points that lie over different manifolds are simultaneously maximized. The LLE based neighborhood reconstruction error for preserving local topological structures leads to accurate low dimensional representation and, hence increases the model’s accuracy. The work in the paper [14] carried out the diffusion on a locally constrained sparse graph to obtain a more accurate graph affinity. A similar kind of work [15] performed the diffusion process on a tensor product graph that captures the higher order information. However, in the aforementioned works, the diffusion process is highly susceptible to noise. This leads to the inaccurate affinity propagation in a graph.
DSCF-net [16] aims to learn an optimal representation matrix along with several sets of basis vectors. The deep factorized coefficient matrix accumulates the similarity information between samples and features. A technique [17] for 2D image is introduced to extract the neighborhood features by minimizing the Frobenius norm-based reconstruction error. This makes the distance metric more reliable and encodes the neighborhood reconstruction error precisely. The RS2ACF [18] framework optimizes the representation by constraining it in both projection and label space. The basis vector obtained from concept factorization is used to discover the latent semantic structure hidden in data proves to be useful for clustering. The label constrained representation matrix preserves the local geometrical information, which propagates accurate labels to unlabeled data. In [19], a parameter free affinity based clustering is introduced that estimate distance between data points, and a threshold affinity is obtained. In paper [20], propose Consistent Affinity Graph Learning (CAGL) algorithm for multi-view data that selects the robust graph Laplacian matrices from each view. It models the hypergraph Laplacians as a point on Grassmann manifold and fuses all the views with CAGL algorithm. In [21], Axiomatic Fuzzy Set-based Spectral Clustering (AFSSC) is proposed that creates a highly robust affinity graph by exploiting and identifying the discriminative features.
Kernel dependency measures like HSIC capture both linear and nonlinear relationships by mapping the data into a high dimensional feature space. A kernel defined on the vertices of a graph gives a representation of the data and measure of similarity between data points in the RKHS. HSIC [22, 23] is calculated from the empirical estimate of the Hilbert-Schmidt norm on the cross-covariance operator. Empirical HSIC is used to measure statistical dependence between two random variables or two datasets assumed to be drawn from different probability density functions. HSIC has been optimized with greedy algorithms on data features [23, 24]. In the paper [25], the proposed feature space independent semi-supervised kernel matching method for domain adaptation. HSIC Lasso using dual augmented Lagrangian for global optimum, has been introduced in [26] for regression with feature reduction. ICA [27], with contrast function based on canonical correlation in the RKHS space, has been proposed for feature selection by finding the minimum correlated dimensions. The HSIC based supervised feature selection [24] measures the dependence between the given features with their respective labels and selects those features that give the maximum correlation. HSIC has also been used for dimensionality reduction [28, 29] such that maximum independent features are identified using the kernel method. The problem of hyperspectral image classification [30] has also utilized surrogate kernel based HSIC to further increase the classification accuracy.
3 Cross-covariance based graph affinity (CCGA)
CCGA based affinity measure has been proposed to capture the nonlinear dependence between data points of a point cloud lying on a Riemannian manifold in place of Euclidean distance. It is recognized that in addition to nonlinear dependence, pairwise affinities are also affected by the presence of common neighbors of data points. The first technique, CCGAL, considers the dependence between neighboring points of a neighborhood to identify innate dependence and to incorporate them in the affinity measure between data points. The second technique, CCGAN assumes that common neighbors do not affect the affinity measure alone but with their own neighborhoods. This effect has been captured in the affinity measure for increased classification accuracy and dimensionality reduction performance.
A pairwise affinity metric needs to capture the mutual dependence (linear or nonlinear) between data points. Semi-supervised learning assumes that the conditional density of the points given the labels varies smoothly along the geodesics in the intrinsic geometry of the marginal distribution of the data points. If two points on the manifold have an edge i.e. high mutual dependence between them on which the marginal density is large, then the two points would generally share the same label. This entails that the affinity metric between two data points must also consider their neighborhoods. Nonlinear dependence can be computed using the kernel in an appropriate feature space. However, to consider the effect of neighborhood requires determination of the neighborhoods of the nodes. This becomes paradoxical as the affinity between data points define edges. Moreover, implicit mapping into the feature space does not preserve neighborhoods. This can be solved by defining local neighborhoods created by k NN or 𝜖 NN using Euclidean distance. This neighborhood is used to determine neighboring points and the effect of neighborhoods on affinity using kernel based dependency measure in the feature space. The resulting affinity is used to find the geodesic on the Riemannian manifold using a heat kernel.
Given n data points \( (x_{i})_{i=1}^{n} \) sampled on manifold \( {\mathscr{M}} \), the local neighborhoods are designed using k-NN or 𝜖-NN represented as G = (X, W) where, X is collection of all data points and W contains the affinity between connected data points. Assume that around each data point xi, the local neighbors are contained in xj ∈ N(xi). The heat kernel based affinity between data points are obtained using:
where, xi is the point of interest, xj ∈ N(xi) is its neighbor, and σ is the heat kernel bandwidth. In graph-based semi-supervised learning methods, affinity measures between labeled and unlabeled data points are used to determine the labels of unlabeled points. The choice of affinity measure is vital in determining the classes of the unlabeled points. A graph is created using k NN such that number of fixed neighbors in k NN may force data points beyond the linear region to be considered as a neighbor. In other situation, genuine linear neighborhood data points may be discarded. Thus, the neighborhood of a data point cannot be guaranteed to hold the local linearity assumption. This requires a non Euclidean measure that can capture the relation between xi and xj ∈ N(xi).
Kernel methods map the input data into a high-dimensional RKHS and define a linear method therein. The model results in a nonlinear relationship with respect to the input space. The mapping is implicit, and the nonlinear relations between data in the feature space are captured through the kernel function. A symmetric positive semi-definite matrix can be viewed as a proximity measure that has an inner product representation and satisfies the criteria required to be a distance metric. Consider for any proximity κ on a finite set X, if two data points xi and xj are connected then the distance between them is obtained using \( d(x_{i},x_{j}) = \frac {1}{2}\left (\kappa (x_{i},x_{i})+\kappa (x_{j},x_{j})-\kappa (x_{i},x_{j})\right ) \). This also indicates that the proximity measures can be combined to generate a distance metric that represents the pairwise affinity or dependence between data points.
In this paper, we follow the proximity approach to encode the similarity among data points through cross covariance based affinity. The method captures both linear and nonlinear properties of the underlying data manifold by means of statistical independence criterion. The cross-covariance operator assigns a value between 0 and 1 where, the former value is assigned for two completely different data points and the later value signifies that the two data points are exactly the same. HSIC of a data point to itself always results in a unity value. As the properties of underlying manifold \( {\mathscr{M}} \) is unknown and the sample data points cannot be guaranteed to have been drawn uniformly. Hence, the distribution parameters of resultant neighborhoods differ from each other. The similarity and differences of neighborhood distribution parameters can only be appropriately utilized when the data is mapped to its full feature space. However, due to the complexity of the mapping and increased dimensions, the kernel proximity automatically identifies the small linear subspace where the manifold properties can be exploited in a linear manner.
The proposed affinity framework relies on defining the local neighborhood on k NN method based on Euclidean distance. This is necessary as the neighborhood is not preserved in the feature space. The implicit kernel mapping redistributes the points. Hence, a Euclidean distance based neighborhood is fixed ab initio. Then the affinities between connected data points are determined through CCGA methods. The proposed techniques account for both linear as well as nonlinear relationships between each pair of connected data points by estimating the HSIC dependence.
3.1 Hilbert-schmidt independence criterion (HSIC)
HSIC is a kernel based similarity measure between two random variables. Given, Q and Z are two random variables sampled from their respective probability density functions. Assume, \( \mathcal {F} \) and \(\mathcal {G} \) are the two RKHSs for Q and Z respectively. Let the ϕ(q) and ψ(z) be the feature mapping functions such that \( \phi (q): Q \rightarrow \mathbb {R} \) and \( \psi (z): Z \rightarrow \mathbb {R} \). The kernel functions are given by \( \kappa (q,q^{\prime }) =\langle \phi (q), \phi (q^{\prime }) \rangle _{\mathcal {F}}\) and \( \hat {\kappa }(z,z^{\prime }) =\langle \phi (z), \phi (z^{\prime }) \rangle _{\mathcal {G}} \). The empirical estimation of HSIC using a finite number of data samples is defined as:
where, n is the total number of observation, κ, and \( \boldsymbol {\hat {\kappa }} \) are the two respective kernel matrix for Q, and Z respectively, and C is the centering matrix. The similarity value between two data vectors in the Hilbert space is given by (2).
3.2 CCGA for local region (CCGAL)
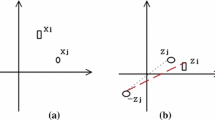
Assume a data point xi and its neighbors xj,⋯ , xk ∈ N(xi). Based on smoothness assumption, if N(xi) is a local region with high affinity between neighborhood data points, then all the neighbors shall share similar label with high probability. This entails that two data points xj ∈ N(xi) and xk ∈ N(xi) that are part of the neighborhood N(xi) of xi but not adjacent to each other i.e. xj∉N(xk) and xk ∈ N(xj) may also generally share the same label. It also indicates that the affinity between such data points xj and xk should be large if they have multiple common neighborhood. However, in general for affinity calculation, the relation between such xj and xk is not considered unless either xj ∈ N(xk) or xk ∈ N(xj) is true. The proposed CCGAL tries to identify such similarities among data points and accumulate them to define revamp affinity between two data points based on two factors, first, whether they are neighbors and second, number of common local neighborhood shared by them.
Figure 1 shows the pictorial representation of CCGAL, xi as point of interest with data points in the k NN neighborhood xj ∈ N(xi). Here, xi is denoted by black filled circle and its neighbors with blue filled circles. The direct connectivity between xi and xj ∈ N(xi) has been illustrated using solid lines which is approximated similarly by both Euclidean distance and CCGAL. This CCGAL determines the statistical independence between the neighbors. This is represented by dashed line. If the data points xj and xk further share common local neighborhood apart from xi, the next such distance should be added to previous value between them. The proximity between any two connected data points xi and xj can be defined as
Since, κ of a data point to itself is 1.
Similarly, when two data points xj, and xj lie in the neighborhood N(xi), then their distance is calculated using
where, \( \kappa ^{(N(x_{i}))}(\cdot ,\cdot ) \) signifies kernel proximity applied the neighborhood of N(xi), and \( w^{(N(x_{i}))} \) represents the respective neighborhood distance. Further, if xj, and xk are immediately connected or are part neighborhoods of other data point (N(⋅)), then the distance between them is obtained by averaging over all individual distances using [31]
The final affinity wjk between any two data points xj and xj where, w(xj, xk)≠ 0 can be obtained using the heat kernel as
The algorithm for CCGA for local region has been explained in Algo. 1.
The CCGAL major steps are as follows,
-
1.
Given a dataset X having n number of points, create a k NN neighborhood N(xi) around each data point xi
-
2.
Apply kernel \( \kappa ^{N(x_{i})} \) and compute the k × k distance matrix.
-
3.
Convert this distance matrix to an affinity matrix as in (5).
-
4.
For each pair of data points xj and xk contained in the current k × k affinity matrix, add the current affinity values to the previously computed values.
-
5.
Repeat the step 2 until all n data points are processed.

3.3 CCGA for neighborhood (CCGAN)
CCGAL is based on the assumption that common neighbors influence the mutual dependence between two data points. These individual influences may be further conditioned by their own neighborhoods. CCGAN tries to assimilate this intangible effect of the neighborhoods of the neighbors themselves in the affinity metric. Again, to adhere to the manifold assumption, a neighborhood is identified using spatial proximity based on Euclidean distance. CCGAN proceeds as follows, given data points in X, a local neighborhood N(xi) can be constructed using k NN or 𝜖 NN around every point xi based on Euclidean distance between all data points. Once the graph is created, we want to compute the pairwise affinity, based on their nonlinear dependence, the effect of neighboring data points, and their respective neighborhoods between the point of interest xi and each of its neighbor xj ∈ N(xi). As shown in Fig. 2, the pairwise affinity between every such pair of xi and xj would further decide if they are likely to share similar labels. We have two neighborhoods N(xi) around the xi and other N(xj) around the xj such that xj ∈ N(xi). To compute the nonlinear dependence, we apply RBF kernels: \( \kappa ^{(N(x_{i}) }\) and \( \kappa ^{(N(x_{j})} \) for N(xi) and N(xj) respectively. Each k NN neighborhood contains k + 1 elements including point of interest. The kernels over N(xi) and N(xj) allows the algorithm to appropriately weigh the importance of neighbors. The HSIC based statistical independence between N(xi) and N(xj) is
w(N(xi), N(xj)), is converted into distance as follows,
The resultant d(N(xi), N(xj)) distance matrix consists of (k + 1) × (k + 1) where each \( d_{i^{\prime }j^{\prime }} \) corresponds to distance between i′th member of N(xi), and \( j^{\prime }_{\text {th}}\) member of N(xj):
The matrix d(N(xi), N(xj)) now contains the distance calculated using HSIC between every pair of N(xi), and N(xj) represented as rows, and columns respectively as shown above. Finally, the distances between data points being member of such N(xi), andN(xj) are summed up together to strengthen such relations. \(w(x_{i},x_{j})=w(N(x_{i}),N(x_{j}))+w(N(x_{i}),N(x_{k}))_{x_{j}\in N(x_{k})}+w(N(x_{j}),N(x_{k}))_{x_{i}\in N(x_{k})}+w(N(x_{k}),N(x_{l}))_{x_{i}\in N(x_{k}),x_{j}\in N(x_{l})}+\dots \)
This distance is calculated between data point and each of its neighbors. Inside every neighborhood, w(N(xi), N(xj)) is computed k times. Over n data points, this algorithm is repeated k × n times to compute the distance matrix. The final affinity wij between any two data points can be obtained using (5). The algorithm for CCGA for local region has been explained in Algorithm 2.

The CCGAN can be summarized in following steps.
-
1.
Given data points in X, local neighborhood N(xi) is obtained with k NN algorithm around each data point.
-
2.
After obtaining this graph, based on the Euclidean the distance, affinity between point of interest xi and each of its neighbor xj ∈ N(xi) is computed. In this way, we have two neighborhoods, N(xi) around xi and N(xj) around xj.
-
3.
On each of this neighborhood, the RBF kernel \( \kappa ^{(N(x_{i}))} \) and \( \kappa ^{(N(x_{j}))} \) is applied.
-
4.
The HSIC based statistical independence between two these neighborhoods is computed as (6).
-
5.
The pairwise HSIC values between data points is converted into distance (7). This forms a distance matrix of size (k + 1) × (k + 1).
-
6.
In every neighborhood w(N(xi), N(xj)) is computed k times which is repeated for n data points (k × n) times.
-
7.
The final affinity matrix is obtained by (3).
3.4 Complexity analysis
The techniques based on CCGA largely depends on the number of data points n and ambient dimension D. For both, CCGAL and CCGAN RBF kernel (\( \mathcal {O}(n) \)) and k NN algorithm (\(\mathcal {O}(D\times n \times k)\)) has been used. The graph mainly depends on the k number of nearest neighbors. The time complexity for both techniques can be expressed as,
-
CCGAL, complexity is computed based on steps as defined in Algorithm 1.
-
1.
Step 5, copying the data points \( \mathcal {O}(1) \).
-
2.
Step 6, number of element count \( \mathcal {O}(k) \).
-
3.
Step 7, matrix subtraction \( \mathcal {O}(k \times d) \).
-
4.
Step 8, identity matrix \( \mathcal {O}(1) \).
-
5.
Step 9, centering matrix \( \mathcal {O}(k \times d) \).
-
6.
Step 10, RBF kernel computation over k data points \( \mathcal {O}(k) \).
-
7.
Step 11, affinity approximation over k data point \( \mathcal {O}(k \times k) \).
The over all time complexity for n data points using CCGAL technique is given as \( \mathcal {O}(n \times k)(d+k) \).
-
1.
-
CCGAN, complexity is computed based on steps as defined in Algorithm 2.
-
1.
Step 5, copying the data points \( \mathcal {O}(1) \).
-
2.
Step 6 & 11, number of element count \( \mathcal {O}(k) \).
-
3.
Step 7 & 12, matrix subtraction \( \mathcal {O}(k \times d) \).
-
4.
Step 13, identity matrix \( \mathcal {O}(1) \)
-
5.
Step 14, centering matrix \( \mathcal {O}(k \times d) \).
-
6.
Step 15 & 16, RBF kernel computation over k data points \( \mathcal {O}(k ) \).
-
7.
Step 17, distance estimation \(\mathcal {O}(k) \)
-
8.
Step 20, conversion into affinity \( \mathcal {O}(k \times k) \)
The over all time complexity for n data points using CCGAN technique is given as \(\mathcal {O}(k^{3} \times n \times d)(d+k) \).
-
1.
4 Experiment and Results
The performance of the proposed graph affinity based techniques CCGAL and CCGAN have been evaluated on both synthetic and real world datasets for manifold learning and graph Laplacian regularized classification, i.e. manifold regularization. The existing state-of-the-art graph affinity methods like heat, binary EMR [32] (both 24 and 72 graphs) used for graph construction are compared against our techniques in terms of classification error. In order to validate the robustness of our techniques, a wide range of real world datasets are used. The real world datasets mainly comprise of images, handwritten characters, scene recognition and brain computer interface. The binary classification error is estimated with two models LapSVM and LapRLSC for both test and unlabeled sets. The mean classification errorFootnote 1, along with standard deviation by varying the value of k are summarized in the tables.
4.1 Synthetic datasets
The synthetic datasets contain 3D manifold shapes whose low dimensional 2D representation is known. Non-linear dimensionality reduction based 2D projections using both CCGAL and CCGAN techniques are obtained. For comparison many existing state-of-the-art non-linear dimensionality reduction techniques namely LTSA (Local Tangent Space Alignment) [33], LLE (Local Linear Embedding) [34], Isomap (Isometric Mapping) [35], LE (Laplacian Eigenmap) [36], and HE (Hessian Eigenmap) [37] are employed along with linear dimensionality reduction technique PCA (Principal Component Analysis) [38]. The first row of figures in the Table 2 contains the original 3D manifolds and subsequent rows contain the 2D output from each of the employed techniques.
-
1.
Punctured Sphere: The second column of the Table 2 shows the punctured sphere dataset. This artificially generated dataset contains 4000 data points in 3D. It is originally a 2D disc, which is elevated on the third dimension to make it a sphere. The results show that PCA, LLE, Isomap and HE are unable to determine the true 2D structure as they failed to preserve the intrinsic geometry of the punctured segment marked with red color. Both CCGAL and CCGAN, along with LTSA and LE preserve the true intrinsic geometry and are able to unfold the true lower space structure. The HSIC based nonlinear dependence is able to find the true affinity among data points which leads to a better association among data points belonging to the same class.
-
2.
Sine on a hyperboloid: The third column of the Table 2, shows 629 data points, sine wave wrapped around a hyperboloid in 3D space which is originally a smooth circle on a 2D plane. As evident, CCGAL, CCGAN, and Isomap give the best representation in 2D space while retaining the geometrical characteristics. All other techniques fail to either preserve the circle or its smoothness. These techniques only rely on the Euclidean distance for structural properties estimation and discard a large chunk of the nonlinear properties.
-
3.
Swiss roll with hole: This dataset contains 3947 data points modeled in a Swiss roll shape with a small hole in between green and blue portion. When unrolled correctly, it should appear as a 2D flat strip with a hole. Isomap is able to give the closest output to its intrinsic representation. LLE, CCGAL, and CCGAN are able to preserve the shape of the hole along with the strip. Other methods failed to preserve the connectivity around the hole. CCGAL, and CCGAN are not able to better unroll the structure. It seems that they discard some spurious dependencies between points in the hole perimeter.
-
4.
Twin peak with hole: The last column of the Table 2, shows the artificially generated Twin peaks with a hole, which is originally a 2D flat strip with a hole containing 6990 data points. The results show that PCA gave the most accurate representations, followed by CCGAL and CCGAN. Other methods either failed to unroll the strip to its original structure or the boundary data points remained cluttered. All nonlinear dimensionality reduction techniques are able to retain the hole, but 2D shape distortion is maximum for Isomap as the shortest is not the correct geodesic distance.
Manifold learning results as shown in the Table 2 on the synthetic datasets clearly suggest that both the techniques are able to determine the optimal affinities that preserve the maximum intrinsic geometrical properties. This leads to a better association among data points belonging to the same class. It is observed that for all the synthetic datasets, both, CCGAL and CCGAN are able to unroll the data and project them to 2D space while retaining the maximum structural characteristics.
4.2 Real world datasets
-
1.
USPS: This dataset [39] contains scanned images (Fig. 3a) of handwritten digits on envelopes of the US postal services. The original scanned digits varying in size and orientation is resized to 16 × 16 grayscale images with 7291 training and 2007 test instances.
-
2.
Hasy_v2: This dataset [40] comprises of 168233 single handwritten latex symbols (Fig. 3b) across 369 different classes extracted from HWRT dataset. However, only 9 symbols have been used in the experiment as they contained ≥ 800 samples. The original data matrix is of size 12701 × 3072.
-
3.
MNIST : This datasetFootnote 2 contains the images (Fig. 3c) of handwritten digits (0 − 9). Thus, it has 10 classes, each corresponds to a different numeric digit. Each of this image is gray scale of size 28 × 28. The dataset has total of 70,000 instances out of 60,000 has been used for the training and rest for testing.
-
4.
BCI HaLT : BCI dataset [41] comprises of brain electroencephalographic (EEG) signals obtained from different imagery part in micro-volt unit. The recorded EEG signals correspond to brain activity of imagining the movement of left leg, right leg, left hand, right hand and tongue. Across multiple subjects, the experiment contained 3 HaLT reading for subject A only of size 2898 × 3570.
-
5.
CIFAR-10 : This dataset [42] comprises of color images (Fig. 3d) of the objects like cars, cats, airplanes, deer, birds, ships, dogs, trucks, horses, and, frogs. It has 10 different classes with a total of 60,000 colored images. Each of this image is of size 32 × 32 with 6000 images per class.
-
6.
Lego Bricks : This datasetFootnote 3 comprises of gray scale images (Fig. 3g) of the bricks manufactured by Lego. It has total of 16 different classes and each class has ≈ 400 images. Each image is of size 200 × 200 grayscale pixels.
-
7.
UC Merced Land : This dataset [43] has outdoor images (Fig. 3e) of land divided into 21 classes. Each of this image has been obtained from the USGS National Map Urban Area Imagery collection for various urban areas around the country. Each of this class has 100 images and each image is of size 256 × 256. Out of these 100 images, 50 images per class are randomly picked for the training set and rest 50 images per class are used for testing set.
-
8.
CVPR’09 : This dataset [44] comprises of 67 classes of image data (Fig. 3f). Each class has more then 100 images. Each image has been reduced to equal size of 32 × 32 × 3. Finally, a matrix of size 15620 × 3072 which is further divided into training 70% and testing set
-
9.
Natural Images : This dataset [45] contains images (Fig. 3h) of different objects like car, cat, airplane, motorbike, flower, person and fruit. It has total of 8 classes and each this grayscale image has been resized to 28 × 28.. The complete dataset is of size 7599 × 784.
-
10.
COVID-19 CT Scan Dataset : This dataset [46] contains CT scan images (Fig. 3i) of COVID-19 positive and negative patients. It has a total of 451 CT scan grayscale images out of which 275 belonged to positive patients and rest 195 were negative. During pre-processing, all images were changed to standard 512 × 512pixels. Data was further split to 50% as training and 50% as testing set. The experiment was repeated 20 times, and in each round, 8 images from both classes were randomly labeled.
Extensive experiments are performed with LapSVM and LapRLSC classifiers. Root mean square error with standard deviation is reported in separate tables for unlabeled and test set Experiments on these datasets are performed by varying the k NN paramete as the affinity matrix used for constructing the graph depends on k, the number of neighbors. This parameter k is varied from 6 to 15. It is observed that the classification accuracy for all datasets increases, thus, with enriched neighborhood more accurate affinity is learned. On further increasing the value (k = 16) the accuracy suddenly dips and similar trend is followed on further increasing the neighborhood. The classification error for value of k = 6 and k = 10 for both LapSVM and LapRLSC are reported in the Tables 3, 4, 5, 6, 7, 8, 9, and 10.
On USPS dataset, CCGAL and CCGAN have highest classification accuracy as compared to the existing state-of-the-art techniques for both test and unlabeled set. On USPS test set, CCGAN achieves the classification accuracy of > 98% and CCGAL stands closest to it with accuracy > 97%. On the other hand, classification accuracy with heat and binary affinities are > 95% and > 94% respectively. In contrast, EMR24 and EMR72 have < 72% classification accuracy, which is least among all. A similar trend in the classification accuracy is obtained for the unlabeled set, where CCGAN has the best accuracy of > 95.5% and EMR72 has the least accuracy of < 67%. This reveals that HSIC based dependence value obtained between data points is able to capture the nonlinear dependence between data points. This leads to better classification accuracy. On HASY_v2 dataset for test as well as unlabeled set CCGAN has most accurate result > 98% followed by CCGAL (> 98%) and binary (> 97%). EMR24 and EMR72 have almost same accuracy (≈ 87%). Heat affinity has the worst classification accuracy ≈ 78% against others. Both LapRLSC and LapSVM have approximately similar result for Hasy_v2 dataset. This result trend suggests that the CCGAN better decodes the neighborhood connectivities to determine a more enriched affinity matrix. On MNIST dataset for test set, CCGAL provides the most accurate result for handwritten digits with classification accuracy ≈ 87% whereas other proposed technique CCGAN has the classification accuracy ≈ 82%. Only accuracy obtained with binary affinity stand closest to our techniques (> 90%). Other techniques have comparatively very poor classification accuracy (< 65%). On other hand for unlabeled set, results are much better for all techniques like CCGAN, CCGAL and binary weight have ≈ 98% accuracy. Again EMR24 and EMR72 have the least accuracy. On BCI HaLT dataset, all techniques have almost same classification accuracy for both test and unlabeled set under LapSVM as well as LapRLSC. Binary weight and CCGAN have the highest accuracy (> 83%). Whereas, others like heat affinity, CCGAL, EMR24 and EMR72 have accuracy that lies between 78% to 80%. This shows that dataset intrinsic geometry has been well explored by the given techniques in order to determine the connectivities by accommodating both linear and nonlinear characteristics. On CIFAR-10 dataset, there is a similar trend in the classification result. For both test and unlabeled sets classification accuracy has not much variation, the accuracy is ≈ 70%. CCGAN leads among all by 1% with classification accuracy 71%. On another object detection dataset, like Lego Bricks, CCGAL and CCGAN give best label prediction results. They achieve an accuracy of > 94% for both test and unlabeled sets. The next most accurate one is binary weight with accuracy > 91%. EMR24 and EMR72 are the worst performer with only > 86% accuracy. These result values indicate that the graphs obtained with EMR24 and EMR72 discard the major chunk of information present in the feature matrix of dataset. On other hand CCGAL and CCGAN better explore the feature matrix to build a high information contained graph. On UC Merced dataset, CCGAN has the highest accuracy (> 77%) under LapSVM and LapRLSC for test set. Whereas EMR24 (> 78%) under LapSVM for unlabeled set and binary weight (72%) for test set under LapRLSC. There is a slight increase in the accuracy for all techniques under both LapSVM and LapRLSC for k = 10. For CVPR’09 dataset, the proposed techniques CCGAL and CCGAN outperform others for both test and unlabeled sets. The average accuracy for all techniques is ≈ 72% under both LapSVM and LapRLSC. On Natural image dataset, CCGAN outperform other techniques by a non negligible factor. Both CCGAL and CCGAN techniques have > 84% classification accuracy for both test and unlabeled set. Other techniques have accuracy ≈ 80% under LapSVM and LapRSLC. For COVID-19 CT dataset, heat affinity has the maximum classification accuracy of ≈ 90% and other techniques have accuracy > 92% for both unlabeled and test sets. The nonlinear dependencies estimated with HSIC determine the connectivities among data points which leads to better association among same class data points.
In order to further validate the efficacy of CCGAL and CCGAN, random walk plots are constructed with state-of-the-art graph affinities on BCI the dataset are shown in the Fig. 4. An affinity matrix is considered to be better than other affinity matrices if its random walk plot has clear clusters. As evident, the random walk is shown in Fig. 4b constructed using CCGAN contains cluster patches that define a strong intra-neighbor connectivity and sparse inter-neighbor edges. This supports the superior performance of CCGAN during classification as the affinity matrix helps in identifying clear clusters on the data points. The random walk using CCGAL (Fig. 4a) gives a comparable result wherein different classes of the dataset are discernible. The plots for EMR24 (Fig. 4c) and EMR72 (Fig. 4d) have less clear clusters. On the other hand, the random walk over non-parametric methods KTAFootnote 4 and HHG4 do not have much recognizable clusters, while the former affinity do define some high strength groups, the later gave uniform affinity to all data points. Similarly, the heat affinity random walk also diluted the distinguishable clusters by assigning high affinity to every edge (Fig. 4e) as it captures linear dependencies only.
It is evident from the results of non-linear dimensionality reduction and the random walk plots that CCGAL and CCGAN are capable of preserving the smooth intrinsic geometrical properties better than other methods. Similarly, increased classification accuracy across the standard handwritten, BCI, object and scene detection dataset show that the graph affinity obtained gives information about the unexplored connectivities in the data which outperform the other affinity methods by a considerable margin.
4.3 Non parametric statistical tests
CCGAL and CCGAN are based on HSIC, which is a non-parametric statistical measure. While in the previous experiments, they have been compared with existing state-of-the-art parametric affinity methods, it is essential to compare them with state-of-the-art non-parametric statistical test measures. We have selected two such methods, namely kernel target alignment (KTA) [47] and Heller Heller Gorfine (HHG) [47]. As KTA works in feature space by aligning two kernel functions or a kernel and a target function which makes it very similar to HSIC based affinity method. On the other hand, HHG computes the statistical measure from the underlying Euclidean metric, which further allows the comparison to explore if statistical test based on Euclidean norm can appropriately enforce the smoothness constraint in the presence of nonlinear relation. Both KTA and HHG aim to compute the dependence between the samples of two random variables assuming they are not independent of each other.
KTA tries to learn data embedding in feature space such that the data points follow nice clustering properties i.e. data points belonging to the same class should be mapped spatially closer to each other as compared to data points from different classes. Thus, the intra-class distance between data points in feature space remains close to zero, and inter-class distance, greater than zero. Over the data points in X, KTA employs two kernels \( \boldsymbol {\kappa }_{1}={\sum }_{i=j=1}^{n}\kappa _{1}(x_{i},x_{j}) \) and \( \boldsymbol {\kappa }_{2}={\sum }_{i=j=1}^{n}\kappa _{2}(x_{i},x_{j}) \). Further, the alignment between κ1 and κ2 is performed through
where, \( \langle \boldsymbol {\kappa }_{1},\boldsymbol {\kappa }_{2}\rangle _{\mathtt {F}}={\sum }_{i=j=1}^{n}\kappa _{1}(x_{i},x_{j})\kappa _{2}(x_{i},x_{j}) \). It is also interpreted as the cosine angle between two bi-dimensional vectors κ1 and κ2. The measure of alignment between two different kernels on same pair of data points allows to identify true linear neighbors situated in feature space, this corresponds to small spatial distance or larger affinity. Similarly, the non-aligning kernels of data points would result in smaller affinity values representing absence of linear relationship as required by regularization to enforce function smoothness across the dense linear regions.
The other non-parametric statistical test HHG estimates the dependence between two random variables through norm distance. HHG assumes that if the two random variables X and \( X^{\prime } \) are dependent then there exists a point \( (x_{0},x^{\prime }_{0}) \) and radii Rx and \( R_{x^{\prime }} \) such that the joint distribution of X and \( X^{\prime } \) should be different from product of the marginal distribution of balls around \( (x_{0},x_{0}^{\prime }) \). The independence test performed with HHG is given by,
where, S(i, j) is computed by observing the two dichotomous random variables for n observation in both random variables,
Both KTA and HHG values have been used as an affinity metric in place of CCGAN. Further, LapSVM and LapRLSC were executed for both methods on all benchmark datasets. The performance of all three non-parametric affinity methods have been compared on the mean classification error over both unlabeled and test set. The mean error for test and unlabeled set have been reported in Table 11 and Table 12. On USPS dataset, for both test and unlabeled set, CCGAN outperformed both KTA and HHG by achieving an accuracy of > 98%. KTA also gave a close result and remained at second position whereas HHG gave poor accuracy for LapSVM (> 71%). In case of Hasy_v2 dataset, result of all three non-parametric affinity methods remained close and CCGAN managed to marginally outperform others. A similar trend can be observed in other datasets like MNIST, BCI HaLT, CIFAR-10 and Lego brick dataset. Though for all these datasets, the classification accuracy of CCGAN remained only marginally better than KTA and HHG. In remaining datasets, out of the three non-parametric affinity methods, HHG gave the least accurate results whereas the accuracy results of KTA and CCGAN remained very close.
The non parametric statistical evaluation carried out on various datasets in terms of classification accuracy suggests that HSIC based affinity gave more accurate intrinsic information. From Tables 11 and 12, it can be concluded that KTA and CCGAN are performed similar in classification accuracy and same trend has been repeated for all datasets. However, HHG based error values remained very high in comparison to both KTA and CCGAN. This reason for this poor performance of HHG is the Euclidean distance based metric. As this Euclidean distance, which proves to be inefficient in encoding the relations between the data points accurately.
5 Conclusion
In this paper, we proposed Cross-Covariance based Graph Affinity between two data points that incorporates nonlinear dependence, the influence of neighboring data points, and the neighborhoods around data points. The first technique CCGAL, tried to include the influence of the data points in the common neighborhood in the affinity metric between two points. The second technique CCGAN, took into consideration the influence of the neighborhoods of data points that are adjacent to both the points. Both these techniques were able to determine the linear and nonlinear affinity between each data point. The classification accuracy and dimensionality reduction on both real world and synthetic datasets clearly suggested that the influence of adjacent points must be considered in pairwise affinity. The classification accuracy on different categories of datasets like Object detection, handwritten, and scene detection was found to be consistently better in comparison to other methods by a margin of ≈ 1% to 5% under both LapSVM and LapRLSC. CCGAN, has better classification accuracy and dimensionality reduction results that suggest that nonlinear dependence and neighborhoods have significant influence on affinity between data points.
Notes
For explanation refer Section 4.3.
References
Zelnik-Manor L, Perona P (2005) Self-tuning spectral clustering. In: Advances in neural information processing systems, pp 1601–1608
Bach FR, Jordan MI (2006) Learning spectral clustering, with application to speech separation. J Mach Learn Res 7:1963–2001
Pavan M, Pelillo M (2006) Dominant sets and pairwise clustering. IEEE transactions on pattern analysis and machine intelligence 29(1):167–172
Premachandran V, Kakarala R (2013) Consensus of k-nns for robust neighborhood selection on graph-based manifolds. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 1594–1601
Nie F, Wang X, Huang H (2014) Clustering and projected clustering with adaptive neighbors. In: Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining, pp 977–986
Zhu X, Change Loy C, Gong S (2014) Constructing robust affinity graphs for spectral clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1450–1457
Aggarwal CC (2007) On density based transforms for uncertain data mining. In: 2007 IEEE 23rd International Conference on Data Engineering, pp 866–875. IEEE
Soleimani BH, Matwin S, De Souza EN (2015) A density-penalized distance measure for clustering. In: Canadian conference on artificial intelligence, pp 238–249. Springer
Azizyan M, Singh A, Wasserman L, et al. (2013) Density-sensitive semisupervised inference. The Annals of Statistics 41(2):751–771
Nicolau M, McDermott J, et al. (2018) Learning neural representations for network anomaly detection. IEEE transactions on cybernetics 49(8):3074–3087
Terziyan V (2017) Social distance metric: from coordinates to neighborhoods. Int J Geogr Inf Sci 31(12):2401–2426
Bai X, Yang X, Latecki LJ, Liu W, Tu Z (2009) Learning context-sensitive shape similarity by graph transduction. IEEE Transactions on Pattern Analysis and Machine Intelligence 32(5):861–874
Zhang Y, Zhang Z, Qin J, Zhang L, Li B, Li F (2018) Semi-supervised local multi-manifold isomap by linear embedding for feature extraction. Pattern Recogn 76:662–678
Yang X, Koknar-Tezel S, Latecki LJ (2009) Locally constrained diffusion process on locally densified distance spaces with applications to shape retrieval. In: 2009 IEEE conference on computer vision and pattern recognition, pp 357–364. IEEE
Yang X, Prasad L, Latecki LJ (2012) Affinity learning with diffusion on tensor product graph. IEEE transactions on pattern analysis and machine intelligence 35(1):28–38
Zhang Y, Zhang Z, Zhang Z, Zhao M, Zhang L, Zha Z, Wang M (2020) Deep self-representative concept factorization network for representation learning. In: Proceedings of the 2020 SIAM international conference on data mining, pp 361–369. SIAM
Zhang Z, Li F, Zhao M, Zhang L, Yan S (2017) Robust neighborhood preserving projection by nuclear/l2, 1-norm regularization for image feature extraction. IEEE Trans Image Process 26(4):1607–1622
Zhang Z, Zhang Y, Liu G, Tang J, Yan S, Wang M (2019) Joint label prediction based semi-supervised adaptive concept factorization for robust data representation. IEEE Trans Knowl Data Eng 32(5):952–970
Mukhoty B, Gupta R, Lakshmanan K, Kumar M (2020) A parameter-free affinity based clustering. Appl Intell, pp 1–14
Jing P, Su Y, Li Z, Nie L (2020) Learning robust affinity graph representation for multi-view clustering. Inf Sci 544:155–167
Li Q, Ren Y, Li L, Liu W (2016) Fuzzy based affinity learning for spectral clustering. Pattern Recogn 60:531–542
Aronszajn N (1950) Theory of reproducing kernels. Transactions of the American mathematical society 68(3):337–404
Song L, Smola A, Gretton A, Bedo J, Borgwardt K (2012) Feature selection via dependence maximization. The Journal of Machine Learning Research 13(1):1393–1434
Fukumizu K, Bach FR, Jordan MI (2004) Dimensionality reduction for supervised learning with reproducing kernel hilbert spaces. J Mach Learn Res 5(Jan):73–99
Xiao M, Guo Y (2014) Feature space independent semi-supervised domain adaptation via kernel matching. IEEE transactions on pattern analysis and machine intelligence 37(1):54–66
Masaeli M, Fung G, Dy JG (2010) From transformation-based dimensionality reduction to feature selection
Bach FR, Jordan MI (2002) Kernel independent component analysis. Journal of machine learning research 3:1–48
Chen J, Stern M, Wainwright MJ, Jordan MI (2017) Kernel feature selection via conditional covariance minimization. In: Advances in Neural Information Processing Systems, pp 6946–6955
Zheng X, Ma Z, Li L (2019) Local tangent space alignment based on hilbert–schmidt independence criterion regularization. Pattern Anal Applic, pp 1–14
Damodaran BB, Courty N, Lefèvre S (2017) Sparse hilbert schmidt independence criterion and surrogate-kernel-based feature selection for hyperspectral image classification. IEEE Trans Geosci Remote Sens 55(4):2385–2398
Chebotarev PY, Shamis EV (2005) On a duality between metrics and σ-proximities. arXiv preprint math0508183
Geng B, Tao D, Xu C, Yang L, Hua X-S (2012) Ensemble manifold regularization. IEEE Transactions on Pattern Analysis and Machine Intelligence 34(6):1227–1233
Zhang T, Yang J, Zhao D, Ge X (2007) Linear local tangent space alignment and application to face recognition. Neurocomputing 70(7-9):1547–1553
Roweis ST, Saul LK (2000) Nonlinear dimensionality reduction by locally linear embedding. science 290(5500):2323–2326
Silva VD, Tenenbaum JB (2003) Global versus local methods in nonlinear dimensionality reduction. In: Advances in neural information processing systems, pp 721–728
Belkin M, Niyogi P (2002) Laplacian eigenmaps and spectral techniques for embedding and clustering. In: Advances in neural information processing systems, pp 585–591
Donoho DL, Grimes C (2003) Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data. Proceedings of the National Academy of Sciences 100(10):5591–5596
Wold S, Esbensen K, Geladi P (1987) Principal component analysis. Chemometrics and intelligent laboratory systems 2(1-3):37–52
Hull JJ (1994) A database for handwritten text recognition research. IEEE Transactions on pattern analysis and machine intelligence 16(5):550–554
Thoma M (2017) The hasyv2 dataset. preprint arXiv:1701.08380
Kaya M, Binli M K, Ozbay E, Yanar H, Mishchenko Y (2018) A large electroencephalographic motor imagery dataset for electroencephalographic brain computer interfaces. Scientific data 5:180211
Krizhevsky A, Hinton G, et al. (2009) Learning multiple layers of features from tiny images. Citeseer
Yang Y, Newsam S (2010) Bag-of-visual-words and spatial extensions for land-use classification. In: Proceedings of the 18th SIGSPATIAL international conference on advances in geographic information systems, pp 270–279. ACM
Quattoni A, Torralba A (2009) Recognizing indoor scenes. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp 413–420. IEEE
Roy P, Ghosh S, Bhattacharya S, Pal U (2018) Effects of degradations on deep neural network architectures. arXiv preprint arXiv:1807.10108
Zhao J, Zhang Y, He X, Xie P (2020) Covid-ct-dataset: a ct scan dataset about covid-19. arXiv preprint arXiv:2003.13865
Cristianini N, Shawe-Taylor J, Elisseeff A, Kandola J S (2002) On kernel-target alignment. In: Advances in neural information processing systems, pp 367–373
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yadav, R.K., Abhishek, Verma, S. et al. Cross-covariance based affinity for graphs. Appl Intell 51, 3844–3864 (2021). https://doi.org/10.1007/s10489-020-01986-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-020-01986-9