Abstract
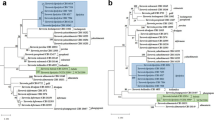
Lipomyces starkeyi is an oleaginous yeast, and has been classified in four distinct groups, i.e., sensu stricto and custers α, β, and γ. Recently, L. starkeyi clusters α, β, and γ were recognized independent species, Lipomyces mesembrius, Lipomyces doorenjongii, and Lipomyces kockii, respectively. In this study, we investigated phylogenetic relationships within L. starkeyi, including 18 Japanese wild strains, and its related species, based on internal transcribed spacer sequences and evaluated biochemical characters which reflected the phylogenetic tree. Phylogenetic analysis showed that most of Japanese wild strains formed one clade and this clade is more closely related to L. starkeyi s.s. clade including one Japanese wild strain than other clades. Only three Japanese wild strains were genetically distinct from L. starkeyi. Lipomyces mesembrius and L. doorenjongii shared one clade, while L. kockii was genetically distinct from the other three species. Strains in L. starkeyi s.s. clade converted six sugars, d-glucose, d-xylose, l-arabinose, d-galactose, d-mannose, and d-cellobiose to produce high total lipid yields. The Japanese wild strains in subclades B, C, and D converted d-glucose, d-galactose, and d-mannose to produce high total lipid yields. Lipomyces mesembrius was divided into two subclades. Lipomyces mesembrius CBS 7737 converted d-xylose, l-arabinose, d-galactose, and d-cellobiose, while the other L. mesembrius strains did not. Lipomyces doorenjongii converted all the sugars except d-cellobiose. In comparison to L. starkeyi, L. mesembrius, and L. doorenjongii, L. kockii produced higher total lipid yields from d-glucose, d-galactose, and d-mannose. The type of sugar converted depended on the subclade classification elucidated in this study.



Similar content being viewed by others
References
Adachi J, Hasegawa M (1996) MOLPHY: Programs for Molecular Phylogenetics ver. 2.3, Computer Science Monographs, No. 28. Institute of Statistical Mathematics, Tokyo
Akaike H (1974) A new look at the statistical model identification. IEEE Trans. Automat Contr AC- 19:716–723
Angerbauer C, Siebenhofer M, Mittelbach M, Guebitz GM (2008) Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresour Technol 99:3051–3056
Efron B (1979) Bootstrap methods: another look at the jackknife. Ann Stat 7:1–26
Fall R, Phelps P, Spindler D (1984) Bioconversion of xylan to triglycerides by oil-rich yeasts. Appl Environ Microbiol 47:1130–1134
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gouliamova DE, Hennebert GL, Smith MT, van der Walt JP (1998) Diversity and affinities among species and strains of Lipomyces. Antonie van Leeuwenhoek 74:283–291
Hasegawa M, Kishino H, Yano K (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Kawahata M, Fujii T, Iefuji H (2007) Intraspecis diversity of the industrial yeast strains Saccharomyces cerevisiae and Saccharomyces pastorianus based on analysis of the sequences of the internal transcribed spacer (ITS) regions and the D1/D2 region of 26S rDNA. Biosci Biotechnol Biochem 71:1616–1620
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kurtzman CP, Albertyn J, Basehoar-Powers E (2007) Multigene phylogenetic analysis of the Lipomycetaceae and the proposed transfer of Zygozyma species to Lipomyces and babjevia anomala to Dipodascopsis. FEMS Yeast Res 7:1027–1034
Lodder J, Kreger-van Rij NJW (1952) The Yeast a taxonomic study. North-Holland Publ. Co., Amsterdam, p 669
Meng X, Yang J, Xu X, Zhang L, Nie Q, Xian M (2009) Biodiesel production from oleaginous microorganisms. Renew Energy 34:1–5
Naganuma T, Uzuka Y, Tanaka K, Koga T (1975) Effect of iron concentration in growth media on growth and development of intracellular oil globule of Lipomyces starkeyi (Factors affecting development of the intracellular oil globule in the yeast Lipomyces starkeyi part I). Nippon Nogeikagaku Kaishi 49:335–340 (in Japanese)
Naganuma T, Uzuka Y, Tanaka K (1985a) Medium for enhancing lipid accumulation and cell proliferation of Lipomyces starkeyi. Nippon Nogeikagaku Kaishi 59:1263–1266 (in Japanese)
Naganuma T, Uzuka Y, Tanaka K (1985b) Physiological factors affecting total cellnumber and lipid content of the yeast, Lipomyces starkeyi. J Gen Appl Microbiol 31:29–37
Naganuma T, Uzuka Y, Tanaka K (1986) Using inorganic elements to control cell growth and lipid accumulation in Lipomyces starkeyi. J Gen Appl Microbiol 32:417–424
Naganuma T, Katsumata K, Ando T, Watanabe H, Nishimura K, Uzuka Y (1999) An improved method for isolating yeasts in the genus Lipomyces and related genera from soil. Biosci Biotechnol Biochem 63:193–195
Nylander JAA (2004) MrModeltest v2 Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smith MT, Kurtzman CP (2011) Lipomyces Lodder & Kreger-van Rij (1952). In: Kurtzman CP, Fell JW, Boekhout (ed) The yeasts, a taxonomic study, 5th edn. Elsevier, pp. 545–547
Smith MT, Poot GA, Batenburg-van der Vegte WH, van der Walt JP (1995) Species delimitation in the genus Lipomyces by nuclear genome comparison. Antonie van Leeuwenhoek 68:75–87
Starkey RL (1946) Lipid production by a soil yeast. J Bacteriol 51:33–50
Suutari M, Priha P, Laakso S (1993) Temperature shift in regulation of lipids accumulated by Lipomyces starkeyi. J Am Oil Chem Soc 70:891–894
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive sequence alignment through sequence weighting, position, specific gap penalties and weight matrix choice. Nuc Acids Res 22:4673–4680
Van der Walt JP, Botha A, Smith MT (1997) Lipomyces mesembrius sp. nov., a member of the L. starkeyi species-complex. Antonie van Leeuwenhoek 71:325–328
Van der Walt JP, Smith MT, Roeijmans HJ (1999) Four new species in Lipomyces. Syst Appl Microbiol 22:229–236
Yang Z (1997) PAML: a program package for phylogenetic and analysis by maximum likelihood. CABIOS 13:555–556
Zhao X, Kong X, Hua Y, Feng B, Zhao Z (2008) Medium optimization for lipid production through co-fermentation of glucose and xylose by the oleaginous yeast Lipomyces starkeyi. Eur J Lipid Sci Technol 110:405–412
Acknowledgments
We thank Mr. Naoto Wakamura for his technical assistance. This study was supported by the New Energy and Technology Development Organization (NEDO), Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2011_9641_MOESM1_ESM.ppt
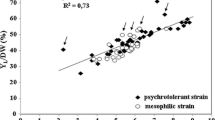
Fig. S1 in supplementary material Scatter diagram of first two components from a principal component analysis with 12 biochemical characters. L. starkeyi s.s. clades = open circle, wild strains in subclade B = open square, wilds strains in subclade C = solid squere, wild strains in subclade D = solid circle, L. mesembrius CBS 7661T, CBS 7600, and CBS 7601 = open diamond, L. mesembrius CBS 7737 = solid diamond, L. doorenjongii = open triangle, L. kockii = cross. (PPT 266 kb)
Rights and permissions
About this article
Cite this article
Oguri, E., Masaki, K., Naganuma, T. et al. Phylogenetic and biochemical characterization of the oil-producing yeast Lipomyces starkeyi . Antonie van Leeuwenhoek 101, 359–368 (2012). https://doi.org/10.1007/s10482-011-9641-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-011-9641-7