Abstract
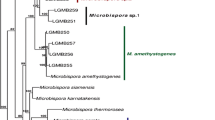
Partial gyrB sequences (>1 kb) were obtained from 34 type strains of the genus Amycolatopsis. Phylogenetic trees were constructed to determine the effectiveness of using this gene to predict taxonomic relationships within the genus. The use of gyrB sequence analysis as an alternative to DNA–DNA hybridization was also assessed for distinguishing closely related species. The gyrB based phylogeny mostly confirmed the conventional 16S rRNA gene-based phylogeny and thus provides additional support for certain of these 16S rRNA gene-based phylogenetic groupings. Although pairwise gyrB sequence similarity cannot be used to predict the DNA relatedness between type strains, the gyrB genetic distance can be used as a means to assess quickly whether an isolate is likely to represent a new species in the genus Amycolatopsis. In particular a genetic distance of >0.02 between two Amycolatopsis strains (based on a 315 bp variable region of the gyrB gene) is proposed to provide a good indication that they belong to different species (and that polyphasic taxonomic characterization of the unknown strain is worth undertaking).




Similar content being viewed by others
References
Al-Musallam AA, Al-Zarban SS, Fasasi YA, Kroppenstedt RM, Stackebrandt E (2003) Amycolatopsis keratiniphila sp. nov., a novel keratinolytic soil actinomycete from Kuwait. Int J Syst Evol Microbiol 53:871–874. doi:10.1099/ijs.0.02515-0
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Bala S, Khanna R, Dadhwal M, Prabagaran SR, Shivaji S, Cullum J et al (2004) Reclassification of Amycolatopsis mediterranei DSM 46095 as Amycolatopsis rifamycinica sp. nov. Int J Syst Evol Microbiol 54:1145–1149. doi:10.1099/ijs.0.02901-0
Chimara E, Ferazoli L, Leão SC (2004) Mycobacterium tuberculosis complex differentiation using gyrB-restriction fragment length polymorphism analysis. Mem Inst Oswaldo Cruz 99:745–748
Chun J, Kim SB, Oh YK, Seong C-N, Lee D-H, Bae KS et al (1999) Amycolatopsis thermoflava sp. nov, a novel soil actinomycetes from Hainan Island, China. Int J Syst Evol Microbiol 49:1369–1373
Coenye T, Gevers D, Van de Peer Y, Vandamme P, Swings J (2005) Towards a prokaryotic genomic taxonomy. FEMS Microbiol Rev 29:147–167. doi:10.1016/j.femsre.2004.11.004
Ding L, Hirose T, Yokota A (2007) Amycolatopsis echigonensis sp. nov. and Amycolatopsis niigatensis sp. nov, novel actinomycetes isolated from filtration substrate. Int J Syst Evol Microbiol 57:1747–1751
Everest GJ, Meyers PR (2008) Kribbella hippodromi sp. nov., isolated from soil from a racecourse in South Africa. Int J Syst Evol Microbiol 58:443–446. doi:10.1099/ijs.0.65278-0
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. doi:10.1007/BF01734359
Jin J, Haga T, Shinjo T, Goto Y (2004) Phylogenetic analysis of Fusobacterium necrophorum, Fusobacterium varium and Fusobacterium nucleatum based on gyrB gene sequences. J Vet Med Sci 66:1243–1245. doi:10.1292/jvms.66.1243
Kasai H, Tamura T, Harayama S (2000) Intrageneric relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int J Syst Evol Microbiol 50:127–134
Keswani J, Whitman WB (2001) Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int J Syst Evol Microbiol 51:667–678
Kim B, Sahin N, Tan GYA, Zakrzewska-Czerwinska J, Goodfellow M (2002) Amycolatopsis eurytherma sp. nov., a thermophilic actinomycetes isolated form soil. Int J Syst Evol Microbiol 52:889–894
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi:10.1007/BF01731581
Labeda DP (1995) Amycolatopsis coloradensis sp. nov., the avoparcin (LL-AV290)-producing strain. Int J Syst Bacteriol 45:124–127
Lechevalier MP, Prauser H, Labeda DP, Ruan JS (1986) Two new genera of nocardioform actinomycetes: Amycolata gen nov. and Amycolatopsis gen nov. Int J Syst Bacteriol 36:29–37
le Roes M, Goodwin CM, Meyers PR (2008) Gordonia lacunae sp. nov. isolated from an estuary. Syst Appl Microbiol. doi:10.1016/j.syapm.2007.10.001
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sensi P, Greco AM, Ballotta R (1959) Rifomycin I isolation and properties of rifomycin B and rifomycin complex. Antibiot Annu 7:262–270
Shen F-T, Lu H-L, Lin J-L, Huang W-S, Arun AB, Young C-C (2006) Phylogenetic analysis of members of the metabolically diverse genus Gordonia based on proteins encoding the gyrB gene. Res Microbiol 157:367–375. doi:10.1016/j.resmic.2005.09.007
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Stackebrandt E, Frederiksen W, Garrity GM et al (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi:10.1099/ijs.0.02360-0
Stackebrandt E, Kroppenstedt RM, Wink J, Schumann P (2004) Reclassification of Amycolatopsis orientalis subsp. lurida Lechevalier et al. 1986 as Amycolatopsis lurida sp. nov., comb nov. Int J Syst Evol Microbiol 54:267–268. doi:10.1099/ijs.0.02937-0
Takahashi K, Nei M (2000) Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol Biol Evol 17:1251–1258
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Tan GY, Robinson S, Lacey E, Brown R, Kim W, Goodfellow M (2007) Amycolatopsis regifaucium sp. nov., a novel actinomycete that produces kigamicins. Int J Syst Evol Microbiol 57:2562–2567. doi:10.1099/ijs.0.64974-0
Thomson CJ, Power E, Ruebsamen-Waigmann H, Labischinski H (2004) Antibacterial research and development in the 21st Century—an industry perspective of the challenges. Curr Opin Microbiol 7:445–450. doi:10.1016/j.mib.2004.08.009
Watanabe K, Nelson JS, Harayama S, Kasai H (2001) ICB database: the gyrB database for identification and classification of bacteria. Nucleic Acids Res 29:344–345. doi:10.1093/nar/29.1.344
Wayne LG, Brenner DJ, Colwell RR et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wink J, Kroppenstedt RM, Ganguli BM, Nadkarni SR, Schumann P, Seibert G et al (2003) Three new antibiotic producing species of the genus Amycolatopsis, Amycolatopsis balhimycina sp. nov., A. tolypomycina sp. nov., A. vancoresmycina sp. nov., and description of Amycolatopsis keratiniphila subsp. keratiniphila subsp. nov. and A. keratiniphila subsp. nogabecina subsp. nov. Syst Appl Microbiol 26:38–46. doi:10.1078/072320203322337290
Wink J, Gandhi J, Kroppenstedt RM, Seibert G, Straubler B, Schumann P et al (2004) Amycolatopsis decaplanina sp. nov., a novel member of the genus with unusual morphology. Int J Syst Evol Microbiol 54:235–239. doi:10.1099/ijs.0.02586-0
Zeigler DR (2003) Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol 53:1893–1900. doi:10.1099/ijs.0.02713-0
Acknowledgements
The authors would like to thank Di James for DNA sequencing. G.J.E. holds a grantholder-linked bursary from the National Research Foundation of South Africa (NRF), a Harry Crossley Foundation Postgraduate Scholarship from the University of Cape Town (UCT), a Twamley Postgraduate Bursary (UCT), a KW Johnstone Research Scholarship (UCT) and a Postgraduate Research Associateship (UCT). This work was supported by research grants to P.R.M. from the Medical Research Council of South Africa, the NRF (grant number: 2073133) and the University Research Committee (UCT).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank accession numbers for the gyrB gene sequences obtained in this study are shown in Table 1.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Everest, G.J., Meyers, P.R. The use of gyrB sequence analysis in the phylogeny of the genus Amycolatopsis . Antonie van Leeuwenhoek 95, 1–11 (2009). https://doi.org/10.1007/s10482-008-9280-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-008-9280-9