Abstract
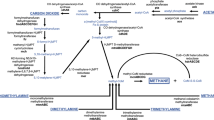
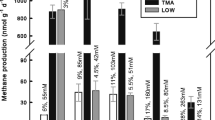
Archaeal microbial communities present in municipal solid waste landfill leachates were characterized using a 16S rDNA approach. Phylogenetic affiliations of 239 partial length 16S rDNA sequences were determined. Sequences belonging to the order Methanosarcinales were dominant in the clone library and 65% of the clones belonged to the strictly acetoclastic methanogenic family Methanosaetaceae. Sequences affiliated to the metabolically versatile family Methanosarcinaceae represented 18% of the retrieved sequences. Members of the hydrogenotrophic order Methanomicrobiales were also recovered in limited numbers, especially sequences affiliated to the genera Methanoculleus and Methanofollis. Eleven euryarchaeal and thirteen crenarchaeal sequences (i.e. 10%) were distantly related to any hitherto cultivated microorganisms, showing that archaeal diversity within the investigated samples was limited. Lab-scale incubations were performed with leachates mixed with several methanogenic precursors (acetate, hydrogen, formate, methanol, methylamine). Microbial populations were followed using group specific 16S rRNA targeted fluorescent oligonucleotidic probes. During the incubations with acetate, acetoclastic methanogenesis was rapidly induced and led to the dominance of archaea hybridizing with probe MS1414 which indicates their affiliation to the family Methanosarcinaceae. Hydrogen and formate addition induced an important acetate synthesis resulting from the onset of homoacetogenic metabolism. In these incubations, species belonging to the family Methanosarcinaceae (hybridizing with probe MS1414) and the order Methanomicrobiales (hybridizing with probe EURY496) were dominant. Homoacetogenesis was also recorded for incubations with methanol and methylamines. In the methanol experiment, acetoclastic methanogenesis took place and archaea hybridizing with probe MS821 (specific for Methanosarcina spp.) were observed to be the dominant population. These results confirm that acetoclastic methanogenesis performed by the members of the order Methanosarcinales is predominant over the hydrogenotrophic and methylotrophic pathways in landfill leachates.
Similar content being viewed by others
Abbreviations
- CLSM:
-
Confocal Laser Scanning Microscopy
- FISH:
-
Fluorescent in situ hybridization
- rDNA:
-
Ribosomal Deoxyribonucleic acid
- rRNA:
-
Ribosomal Ribonucleic acid
References
Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., and Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402
Amann R.I. (1995). In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (eds), Molecular Microbial Ecology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 33.6/1–3.3.6/15
Amann R.I., Krumholz L., and Stahl D.A. (1990). Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172, 762–770
Amann R.I., Ludwig W., and Schleifer K.-H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169
Burggraf S., Mayer T., Amann R., Schadhauser S., Woese C.R., and Stetter K.O. (1994). Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 60, 3112–3119
Chen A.C., Imachi H., Sekiguchi Y., Ohashi A., and Harada H. (2003a). Archaeal community compositions at different depths (up to 30 m) of a municipal solid waste landfill in Taiwan as revealed by 16S rDNA cloning analyses. Biotechnol. Lett. 25, 719–724
Chen A.C., Ohashi A., and Harada H. (2003b). Acetate synthesis from H-2/CO2 in simulated and actual landfill samples. Environ. Technol. 24, 435–443
Chen A.C., Ueda K., Sekiguchi Y., Ohashi A., and Harada H. (2003c). Molecular detection and direct enumeration of methanogenic Archaea and methanotrophic Bacteria in domestic solid waste landfill soils. Biotechnol. Lett. 25, 1563–1569
Daims H., Bruhl A., Amann R., Schleifer K.H., and Wagner M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444
Diaz L.F. (1999). Landfill bioreactor design & operation. Waste Manage. Res. 17, 246–247
Drake H.L., Kusel K., and Matthies C. (2002). Ecological consequences of the phylogenetic and physiological diversities of acetogens. Antonie Van Leeuwenhoek International Int. J. General Mol. Microbiol. 81, 203–213
European Environment Agency E. 2003. Europe’s Environment: The Third Assessment, European Environment Agency, Copenhagen, p. 344
Garcia J.L., Patel B.K.C., and Ollivier B. (2000). Taxonomic phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 6, 205–226
Huang L.N., Chen Y.Q., Zhou H., Luo S., Lan C.Y., and Qu L.H. (2003). Characterization of methanogenic Archaea in the leachate of a closed municipal solid waste landfill. FEMS Microbiol. Ecol. 46, 171–177
Huang L.N., Zhou H., Chen Y.Q., Luo S., Lan C.Y., and Qu L.H. (2002). Diversity and structure of the archaeal community in the leachate of a full-scale recirculating landfill as examined by direct 16S rRNA gene sequence retrieval. FEMS Microbiol. Lett. 214, 235–240
Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., and Stetter K.O. (2002). A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417, 63–67
Hugenholtz P., Goebel B.M., and Pace N.R. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774
Jurgens G., Glockner F.O., Amann R., Saano A., Montonen L., Likolammi M., and Munster U. (2000). Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34, 45–56
Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., Forster W., Brettske I., Gerber S., Ginhart A.W., Gross O., Grumann S., Hermann S., Jost R., Konig A., Liss T., Lussmann R., May M., Nonhoff B., Reichel B., Strehlow R., Stamatakis A., Stuckmann N., Vilbig A., Lenke M., Ludwig T., Bode A., and Schleifer K.H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371
Maidak B.L., Cole J.R., Parker C.T., Jr., Garrity G.M., Larsen N., Li B., Lilburn T.G., McCaughey M.J., Olsen G.J., Overbeek R., Pramanik S., Schmidt T.M., Tiedje J.M., and Woese C.R. (1999). A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27, 171–173
Manz W., Amann R., Ludwig W., Wagner M., and Schleifer K.H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. System. Appl. Microbiol. 15, 593–600
Mori K., Sparling R., Hatsu M., and Takamizawa K. (2003). Quantification and diversity of the archaeal community in a landfill site. Can. J. Microbiol. 49, 28–36
Pohland F.G. (1980). Leachate recycle as a landfill management option. J. Environ. Eng. 106, 1057–1069
Raskin L., Stromley J.M., Rittmann B.E., and Stahl D.A. (1994). Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60, 1232–1240
Reinhart D.R., McCreanor P.T., and Townsend T. (2002). The bioreactor landfill: Its status and future. Waste Manage. Res. 20, 172–186
Stahl D.A. and Amann R. (1991). Development and application of nucleic acid probes. In: Stackebrandt E. and Goodfellow M. (eds), Nucleic Acid Techniques in Bacterial Systematics, pp. 205–248
Uz I., Rasche M.E., Townsend T., Ogram A.V., and Lindner A.S. (2003). Characterization of methanogenic and methanotrophic assemblages in landfill samples. Proc. R. Soc. Lond. Ser. B-Biol. Sci. 270, S202–S205
Wintzingerode F.v., Göbel U.B., and Stackebrandt E. (1997). Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. Fems Microbiol. Rev. 21, 213–229
Acknowledgements
This work was conducted on the MIMOSE experimental platform and was funded by Région Ile de France. The authors would like to thank Nancy Mailly for her excellent technical assistance as well as the two landfill managers who helped us to sample leachate.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Laloui-Carpentier, W., Li, T., Vigneron, V. et al. Methanogenic diversity and activity in municipal solid waste landfill leachates. Antonie Van Leeuwenhoek 89, 423–434 (2006). https://doi.org/10.1007/s10482-005-9051-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-005-9051-9