Abstract
Locating functionally important protein surfaces and identifying the catalytic site residues are critical for studying enzyme functions. Here, we present a method for predicting and characterizing catalytic sites of enzymes that is fold-independent. By extract atomic patterns of catalytic residues in surface pockets computed geometrically, we develop a library of atomic patterns on protein functional surfaces of ca 700 structures. Together with propensities of secondary structures and residue occurrence in active sites, we develop a method to identify functionally important surfaces on protein structures and to locate key residues. We discuss application of our methods to amylase, dioxygenase, deaminase, dehalogenase, and hydratase. A large scale cross-validated prediction study shows that our method is sensitive and specific. Our method can used to study enzyme function, drug design, and engineering novel biochemical function.
Similar content being viewed by others
Introduction
Identifying protein residues that play functional roles is an important task. Proteins have a large number (100–1000) of residues, but only a small fraction of them are directly involved in biochemical functions. These residues often are dispersed in primary sequence, but fold spatially together to form a binding or catalytic surface. A subset of them are key residues because they either directly participate in catalysis, or are important for substrate binding.8,17
Although a large number of protein structures in the Protein Data Bank (PDB) are annotated, e.g., with an enzyme commission (E.C) number representing a specific chemical reaction, often such functional information is incomplete: the location of the binding surface is unknown, the identities of the key residues are unclear, and there are well-known examples where the E.C. labels are misleading. As more protein structures are solved in the structural genomics project,6 a large number of structures have unknown functions. Identifying functionally important surfaces and locating key residues would provide important information for further characterizations.5,10,11
In this study, we develop methods for identifying functional surface from a large set of precomputed surfaces. Our method is based on the analysis of bias of functionally important key residues in composition, in secondary structure, and in atomic patterns. We formulate a probabilistic model for predicting whether a residue located in a surface pocket is functionally important. This model is further used to identify whether a precomputed surface is likely to be important for biological functions. Our paper is organized as follows: we first describe our methods and the data set , we then report results of functional site prediction using several enzymes as example. This is followed by a large scale cross-validation study.
Methods
Data Set from PDB Database
We found there are 13,877 protein structures among >30,000 structures in the Pdb databank that are annotated as enzymes and have enzyme commission (E.C.) numbers. However, in many cases there is no information about where the active site is located on the structure and what residues are involved. We use geometric algorithm to compute surface pockets (including buried voids), which are stored in the CastP database.4 We are able to identify a set \({\mathcal{A}}\) of 3,275 proteins whose surface pockets contain one or more annotated residues as recorded either in the pdb or in the SwissProt database. From these, we select a subset \({\mathcal{B}}\) of ≈ 700 structure after further cleaning up by verifying the annotations for each of the key residues, as well as requiring that experimentally measured B-factor exist. Altogether, this final set of ≈ 700 protein structures contain 3,007 annotated residues. We define a functional surface as a surface pocket containing one or more of annotated key residue(s). Fig. 1a shows the size distribution of functional surface pockets in set \({\mathcal{A}}\) . The mean size is 35 residues. Fig. 1b shows that the amino acid residue composition of these functional surfaces is very different from that of full backbone protein sequences.13
The length distribution and unique residue composition of functional surfaces for 3275 proteins with known key residues. (a) Functional surfaces usually consist of 8–200 residues, with the mean value of 35 residues. (b) The amino acid residue composition of functional surfaces on these proteins is different from the composition of sequences used to construct the Jtt model.13 (c) The distribution of the size ratio (defined as \({\frac{\rm length(pocket)}{\rm length(backbone)}}\)). The ratio ranges from 0.1 to 0.3. Proteins commonly have size from 100 to 450 residues. They are most likely to have functional pockets of length from 10 to 80 residues. (d) The mean molecular volume of functional pockets is 1,332.95 Å3. In general, the molecular volume of a functional pocket is less than 5,000 Å3 and it’s length is less than 80 residues.
Characteristics of Enzyme Binding Surfaces
An important property of the functional surface is its size, e.g., measured in the number of residues it contains (Fig. 1a). We also calculated the ratio of the size of functional surface over the total size of the full protein. We found that in general, about 10–30% of all residues on a protein are involved in enzyme function (Fig. 1c), namely, proteins use 10–30% of their residues to form local binding surfaces for catalysis. Another informative attribute of enzyme functional site is the molecular volumes (Fig. 1d). Based on these observations, we select those precomputed surface pockets containing 10–30% of the residues as candidates for prediction of functional surface.
Enzyme functional surfaces have characteristic usage of amino acid residues. Fig. 2 shows the distribution of the 20 amino acids in annotated residues on the 3,275 surface pockets from set \({\mathcal{A}}\). Similar to previous studies,2,14,15 we found that His, Asp, Glu, Ser and Cys account for more than 80% of active site residues in functional pockets. On the other hand, nonpolar residues (e.g., Val, Leu, Pro) are absent. These hydrophobic resides are enriched in protein core for maintaining protein stability, but play little roles in enzyme activities.
Active site residues are mapped to functional pockets and based on annotation in SwissProt and Pdb (17930 pdb entries). His, Asp, Glu, Ser and Cys account for more than 80% of active site residues of functional pockets. In the contrast, Ala, Pro, Val, Leu and Met are completely missed because they are hydrophobic attracted in the core of proteins.
For each annotated residue, we obtain its atomic pattern by listing the atoms that are exposed on the surface wall of the pocket in a consistent order. In addition, the secondary structural environment (e.g., β-sheet, denoted as s, α-helix h, and coil c) of a residue also provides useful information. For example, backbone N and O atoms form H-bonds in α-helix and β-strand and therefore are expected to be less likely to form H-bond involved in the interaction with substrates. Therefore, we also record the secondary structural environment of this residue: h for helix, s for β-sheet, and c for coil. For example, the Gln208 residue in the alpha-amylase structure 1bag (see Fig. 4b) has the following atomic pattern:
From the 3007 annotated key residues on proteins of set \({\mathcal{B}}\) , we obtain 1031 atomic patterns.
Integrated Predictor of Functionally Important Residues
For a residue i located in a surface pocket, because the identity r i of this residue, its secondary structure environment s i , and its atomic pattern a i all provide useful discriminating information for identifying key residues important for enzyme functions, we use the following method to integrate these parameters and calculate the key residue probability \({\mathbb{P}}(i\in \mathcal{K})\) for the i-the residue to be from the set \({\mathcal{K}}\) of key residues:
where \({\pi(s_i|i\in \mathcal{K})}\) the probability of a key residue to be of the secondary structure type s i , \({\pi(r_i|i\in \mathcal{K})}\) is the probability of a key residue to be amino acids type r i , \({\pi(a_i|i\in \mathcal{K})}\) the probability of a key residue to be of the atomic pattern a i , respectively. These are estimated from the \({\mathcal{B}}\) dataset of annontated key residues. For example, the probability \({\pi(a_i|i \in \mathcal{K})}\) is estimated from the occurrence of a specific atomic pattern a i taken by residue i in all annotated key residues from set \({\mathcal{B}}\).
B-factors as a Filter for Atoms
Temperature B-factors or Debye-Waller factors are experimentally measured for atoms in X-ray crystallography and have been used to represent the atomic mobility. Residues exhibiting relatively low B-factors are generally those participating in forming secondary structures, neighboring disulfide bridges, or are involved in ligands binding. Atoms largely exposed to solvent generally experience more fluctuation and exhibit larger B-factors.
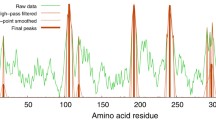
To test the hypothesis whether key resides potentially involved in ligands binding have lower B values, we use ca 500 structures without ligands or substrates from protein set \({\mathcal{B}}\) and compare B-factors of key residues and of non-key resides. Fig. 3 shows that in general key residues have smaller B-factors, and most are polar residues (e.g., His, Asp, Glu, Asn, Gln, Lys, Arg, and Ser).
Based on this observation, we use B-factors as a filter in our predicton. For a surface pocket, we first select only atomic patterns with high probabilities, namely, those appear with high frequencies among all patterns. For atomic patterns with single occurrence that is recorded in the database of known key residues, we compare their B-factors to that of key residues with the same atomic patterns from our database. If the B-factor is less than the highest one from the database, we accept this residue for further analysis, otherwise this residue is removed from further considertion. For multiply occurring patterns, we only accept the ones if their B-factors are less than the average B-factors of the same pattern in the database, or we choose the lowest one. With this implementation of B-factor as a filter, we can improve the accuracy of predicting key residues (Table 1).
Identifying Functional Surface
A functional surface is where protein performs its biological roles. To identify key resides involved in biochemical reactions, a prerequisite is that the functional surface is identified correctly.
We identify the functional surface pocket p from a set of computed pockets \({\mathcal{P}}\) on a protein structure. We compute the summed probability SP(p) for a pocket surface p:
If SP(p) ≥ 10−3, we declare that pocket p is a functional surface.
Results
An Example
We use alpha-amylases 1bag as an example. Alpha-amylase (≈420 residues) acts on starch, glycogen and related polysaccharides and oligosaccharides. Our task is to locate which pocket is the functional surface among the 60 pockets and further identify the key residues involved in the enzymatic reaction. Our only input is the structure of the protein.
We first exhaustively compute all of the pockets (including voids) on this protein structure.4,16 We then compute the key residue probability \({\mathbb{P}(i\in \mathcal{K})}\) for each residues i in a pocket.
We first predict the functional surface. We rank the 60 surface pockets by summed probability SP(p). The largest pocket (CastP ID = 60) contains the largest number (7) of predicted key residues, and has the largest SP(p) = 1.31 × 10−3 value. It is therefore predicted to be the functional surface pocket involved in enzyme reaction. This prediction is correct based on annotation and biochemical literature.
We then predict likely key residues important for enzymatic function after we collect pocket surfaces with SP greater than a threshold θ = 10−3. For this protein structure, pocket 60 is the only one satisfying this condition. It contains 18 residues (Fig. 4). We found that there are four residues whose \({\mathbb{P}(i\in \mathcal{K})}\) values are significantly higher than the rest of 14 residues, and are predicted as key residues. These residues are identical from the annotated residues reported in the literature.7,9
Predicting binding surface and key residues of alpha-amylase. (a) The pocket (green) with Castp ID = 60 is predicted to be the functional surface interacting with the substrate glucose (red). This functional surface contains 18 residues. Four of them are predicted to be functionally important: ASP176 (yellow), HIS180 (cyan), GLN208 (pink) and ASP269 (blue). (b) The four predicted key residues contains several high propensity atomic patterns from our library of 1031 functional atomic patterns. The class of secondary structural environment (β sheet s, helix h , and coil c) is also listed.
Large-scale Prediction of Functional Surfaces
Locating the functional surface is an important task in studying enzyme mechanism, as the correct surface will guide further analysis of binding and catalysis mechanism, and will facilitate the correct prediction of the key residues on protein functional surfaces.12 To evaluate the performance of our method in identifying functional surfaces, we use 10-fold cross-validation tests on the \({\mathcal{B}}\) dataset. We remove 10% of the structures to test the performance of the prediction method, which is derived from the analysis of the rest 90% of the data.
Our results are summarized in a Receiver Operating Characteristics (ROC) curve, where the sensitivity of our method is plotted against its specificity at various significance levels of summed probability values. Here the x-axis represents the false positive rate, namely, 1−specificity, or, 1- TN/(TN+FP), where TN is the number of true negatives, FP the number of false positives. The y-axis represents the true positive rate or sensitivity, defined as TP/(TP+FN), where FN is the number of false negatives.
An overall performance measure is the area under the ROC curve, which is 98.3%, indicating our method performs very well. At the confidence level of summed probability SP = 10−3, the average specificity of our predictions of the functional surfaces of all 3,503 protein surfaces in these 70 proteins in 10-fold cross-validation tests is 99.88%, and the average sensitivity is 92.9% (Fig. 5). Table 2 further provides details of the performace assessed in accuracy (measured as TP/(TP+FP)), with an average value of 91.2%.
Prediction of Key Residues on Protein Functional Surfaces
We compare the predicted key residues with enzymes contained in the Structure-Function Linkage Database (SFLD),18 which links related sequences and structures of enzymes to their chemical reactions, with detailed annotation of enzyme active site residues. We select the four enzyme families that each has 8 or more structures. These are: 2,3-dihydroxybiphenyl dioxygenase (E.C. 1.13.11.39), adenosine deaminase (E.C. 3.5.4.4), 2-haloacid dehalogenase (E.C. 3.8.1.2), and phosphopyruvate hydratase (E.C. 4.2.1.11). We take a random template structure from each protein family, and apply our method to identify functional surfaces and then locating functionally important residues. As shown in Table 3, we are able to accurately locate many functionally important residues.
Conclusions and Future Works
Conclusions
In this work, we have developed a method for identifying functional surfaces and for locating key resides. Our method is sequence and fold independent. We are able to identify systematically functional surfaces with ≥ 91.2% accuracy. In the example of alpha-amylase, functional surface and the key residues identified fully agree with experimental data. Our work provides a fully automated method for locating functionally important surface and for identifying key residues. It can be used to study the mechanism of enzyme reaction, including interactions between residues and substrates. Its applications include drug design and engineered biochemical reactions.
Future Works
We plan to increase the size of the library of annotated functional surfaces, as more structures are being deposited in the Pdb databank. Additional annotations can be incorporated by homology transfer when a surface is matched with another annotated surface satisfying stringent criterion (p-value ≤ 10−5 for cRMSD3 distance of matched surfaces).
We also plan to incorporate evolutionary information in our model. Because residues in protein functional surface experience strong selection pressure,19 we expect this would further improve our method. We plan to further study protein dynamics. Protein function often involves dynamic processes,1 and a crystal structure is only a snapshot conformation of a protein. The shape of the functional surface will change locally and may affect the shape of geometrically computed pockets. We expect that this problem will be alleviated as more structures are deposited and different functional conformations will be increasingly represented in the database. We will examine this issue and assess the robustness of current approach.
References
Bahar, I., Atilgan, A. R., Erman, B. (1997) Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold. Des. 2:173–81
Bartlett, G. J., Porter, C. T., Borkakoti, N., Thornton, J. M. (2002) Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 324:105–121
Binkowski T. A., Adamian L., Liang J. (2003) Inferring functional relationships of proteins from local sequence and spatial surface patterns. J. Mol. Biol. 332:505–526
Binkowski, T. A., Naghibzadeh, S., Liang, J. (2003) CASTp: Computed atlas of surface topography of proteins. Nucleic Acids Res. 31:3352–3355
Binkowski, T. A., Joachimiak, A., Liang, J. (2005) Protein surface analysis for function annotation in high-throughput structural genomics pipeline. Protein Sci. 14:2972–2981
Chandonia, J. M., Brenner, S. E. (2006) The impact of structural genomics: Expectations and outcomes. Science 311(5759):347–351
Collins T., De Vos D., Hoyoux A, Savvides S. N., Gerday C., Van Beeumen J., and G. Feller. Study of the active site residues of a glycoside hydrolase family 8 xylanase. J. Mol. Biol. 354(2):425–435, 2005
Copley, S. D., Novak, W. R., Babbitt, P. C. (2004) Divergence of function in the thioredoxin fold suprafamily: Evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry 43:13981–13995
Fujimoto, Z., Takase, K., Doui, N., Momma, M., Matsumoto, T., Mizuno, H. (1998) Crystal structure of a catalytic-site mutant alpha-amylase from Bacillus subtilis complexed with maltopentaose. J. Mol. Biol. 277:393–407
George, R. A., Spriggs, R. V., Bartlett, G. J., Gutteridge, A., MacArthur, M. W., Porter, C. T., Lazikani, B., Thornton, J. M., Swindells, M. B. (2005) Effective function annotation through catalytic residue conservation. Proc. Natl. Acad. Sci. USA 102:12299–12304
Glaser, F., Morris, R. J., Najmanovich, R. J., Laskowski, R. A., Thornton, J. M. (2006) A method for localizing ligand binding pockets in protein structures. Proteins 62:479–488
Gold, N. D., Jackson, R. M. (2006) Fold independent structural comparisons of protein-ligand binding sites for exploring functional relationships. J. Mol. Biol. 355:1112–1124
Jones, D. T., Taylor, W. R., Thornton, J. M. (1992) The rapid generation of mutation data matrices from protein sequences. CABIOS 8:275–282
Kim, J., Mao, J., Gunner, M. R. (2005) Are acidic and basic groups in buried proteins predicted to be ionized? J. Mol. Biol. 348:1283–1298
Laskowski, R. A., Watson, J. D., Thornton, J. M. (2005) Protein function prediction using local 3D templates. J. Mol. Biol. 351:614–626
Liang, J., Edelsbrunner, H., Woodward, C. (1998) Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 7:1884–1897
Meng, E. C., Polacco, B. J., Babbitt, P. C. (2004) Superfamily active site templates. Proteins 55:962–976
Pegg, S. C., Brown, S. D., Ojha, S., Seffernick, J., Meng, E. C., Morris, J. H., Chang, P. J., Huang, C. C., Ferrin, T. E., Babbitt, P. C. (2006) Leveraging enzyme structure-function relationships for functional inference and experimental design: The structure-function linkage database. Biochemistry 45:2545–2555
Tseng, Y. Y., Liang, J. (2006) Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: A Bayesian Monte Carlo approach. Mol. Biol. Evol. 23:421–436
Acknowledgments
This work is supported by grants from NSF (CAREER DBI0133856), NIH (GM68958), and ONR (N000140310329).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tseng, Y.Y., Liang, J. Predicting Enzyme Functional Surfaces and Locating Key Residues Automatically from Structures. Ann Biomed Eng 35, 1037–1042 (2007). https://doi.org/10.1007/s10439-006-9241-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-006-9241-2