Abstract
Frugivorous bats play a vital role in tropical ecosystems as pollinators and seed dispersers but are also important vectors of zoonotic diseases. Myanmar sits at the intersection of numerous bioregions and contains habitats that are important for many endangered and endemic species. This rapidly developing country also forms a connection between hotspots of emerging human diseases. We deployed Global Positioning System collars to track the movements of 10 Indian flying fox (Pteropus giganteus) in the agricultural landscapes of central Myanmar. We used clustering analysis to identify foraging sites and high-utilization areas. As part of a larger viral surveillance study in bats of Myanmar, we also collected oral and rectal swab samples from 29 bats to test for key emerging viral diseases in this colony. There were no positive results detected for our chosen viruses. We analyzed their foraging movement behavior and evaluated selected foraging sites for their potential as human-wildlife interface sites.
Similar content being viewed by others
Introduction
Wild populations of animals have been well documented to act as reservoirs for zoonotic disease (Kruse et al. 2004). Highly mobile animals such as birds and bats have heightened potential to spread pathogens and study of their movement behavior can shed light on potential pathways for emerging epidemics (Daversa et al. 2017; Feare 2010). Bats are unique as the only true flying mammal, and their capacity to carry and transmit many infectious diseases to other species, including humans, has been widely reported (Hughes et al. 2007; Mackenzie et al. 2003; Olival et al. 2015). Many species of bats are known to host a range of zoonotic viruses, several of which have proven fatal to humans including rabies, Marburg, Nipah, and Hendra-viruses. More recently, bats have been identified as the natural host of several coronaviruses responsible for human fatalities including severe acute respiratory syndrome (SARS-CoV-1) and Middle East respiratory syndrome (MERS-CoV) (Calisher et al. 2006; Machhi et al. 2020). Moreover, bats are largely speculated to be the primary host for SARS-CoV-2, the causative agent for the COVID-19 pandemic (Zhou et al. 2020). Earlier studies have suggested that both ecological and physiological factors may constitute risk factors for infection in bats and potentially play a role in the spillover of viral infections to other species (McMichael et al. 2016). A better understanding of the physiological and behavioral qualities of target populations of bats provides additional tools for predicting and preventing emerging viral epidemics.
Pteropids, including the group of frugivorous bats known as flying foxes, have become a group of great interest for their role in the emergence and re-emergence of novel and known viruses with human health implications, as well as their ecological role in the context of the global decline in bat populations (Hughes et al. 2007; Hutson et al. 2001; Mickleburgh et al. 1992, 2002; Young and Olival 2016). Frugivorous bats play a crucial role in tropical ecosystems as pollinators and seed dispersers (Abedi-Lartey et al. 2016; Aziz et al. 2021; Corlett 2009; Oleksy et al. 2017). They are known to travel long distances to forage and, in some cases, seasonally migrate (Breed et al. 2010; Epstein et al. 2009; Fahr et al. 2015; Welbergen et al. 2020). These bats are under threat from hunting, deforestation, the introduction of exotic predators, and environmental disturbances such as natural disasters and climate change (Allen-Wardell et al. 1998; Hahn et al. 2014). Several species of flying foxes have been identified as a primary reservoir for many of the aforementioned zoonotic viral diseases of global concern including filo-, corona-, and henipa-viruses, among others which may have potentially severe effects on human health (Chong et al. 2009; Mackenzie and Williams 2009). With fruit as their primary food source, this brings flying foxes into regular contact with agricultural plantations, livestock, and humans (Syafiq et al. 2016). As environmental disruption continues to increase as a result of human population growth and agricultural expansion, bats and other wildlife increasingly come into contact with humans, leading to increased human exposure to known and novel pathogens (Giles et al. 2018; Hughes et al. 2007; Plowright et al. 2011). This increased contact may have negative consequences, both for the human populations at risk for disease emergence, as well as for bats and other wildlife under threat of habitat loss.
Myanmar sits at a key juncture between countries and bioregions in Southeast Asia, forming a connection between India, China, Vietnam, Laos, Thailand, and peninsular Malaysia. With 60% of the country still forested, Myanmar provides crucial habitat for many resident and migratory animal species (Bhagwat et al. 2017). While forming an ecological connection between bioregions, Myanmar also connects countries that are known hotspots for emerging human infectious diseases (Jones et al. 2008). With a rapidly growing population and developing tourism and agricultural industries, increased contact with wild populations of animals and increased human and animal movement across borders put Myanmar at risk for emerging zoonotic diseases (McEvoy et al. 2019; Nijman 2010; Nijman and Shepherd 2014; Valitutto et al. 2020).
Many species of frugivorous bats have been reported to reside in Myanmar including Pteropus giganteus, P. hypomelanus, P. intermedius, P. vampyrus, Rosettus leschenaultii, R. amplexicaudatus, Cynopterus sphinx, and C. brachyotis (Bates et al. 2000; Oo et al. 2017; Struebig et al. 2005). Viral metagenomic studies in Myanmar have identified viruses from 24 viral families residing in bats from Myanmar, including the isolation of a hepatitis virus which may have human health implications (He et al. 2013a, 2013b). Transmission of viruses from wildlife, including frugivorous bats, to human populations has been extensively researched (Brierley et al. 2016; Ge et al. 2012; Hasebe and Le 2007; Olival et al. 2013, 2015; Young and Olival 2016). Nucleic acids from viruses including coronaviruses, parvoviruses and filoviruses have been detected in oral, rectal, and urinary samples from many species of bats (Ge et al. 2012; Memish et al. 2013; Schuh et al. 2017). Fresh date palm juice or fruit contaminated with the saliva or fecal material of bats have been implicated in the transmission of Nipah virus infections from bats to humans (Epstein et al. 2008; Hsu et al. 2004; Luby et al. 2006), while shared fruit between fruit bats and pigs has been implicated in the transmission of Nipah virus (Chua 2003) and frugivorous bats have been documented to transmit Henipaviruses via urine, feces, and saliva (Willoughby et al. 2017).
Previous studies have explored the presence of potentially infectious viruses in frugivorous bat populations (Breed et al. 2010; Epstein et al. 2008; Mackenzie et al. 2003; Watanabe et al. 2010), and a number of studies have focused on their movement behavior, navigational capacity and spatial memory (Calderón-Capote et al. 2020; Harten et al. 2020; Sapir et al. 2014; Toledo et al. 2020). However, these two elements have not yet been combined in a single published study where the same bats were sampled for viruses and their movements recorded. Studies on Old World fruit bats, particularly the straw-colored fruit bat (Eidolon helvum), have demonstrated that large bats display species-specific movement preferences in terms of roost habits, range of forage, and seasonal movement variations, all of which may play significant roles in disease ecology and spread of transmissible illness (Epstein et al. 2009; Gumal 2004; Smith et al. 2011; Tidemann and Nelson 2004; van Toor et al. 2019; Welbergen et al. 2020). The lack of data on disease presence and prevalence and the movement dynamics within individual bat populations limits our ability to accurately predict and track emerging viral diseases of concern through bat populations. This lack of knowledge also limits the identification of at-risk human populations, and the capability to halt outbreaks while still residing in the wildlife reservoir. In this study, we sampled Indian flying fox (Pteropus giganteus) for a range of potential zoonotic diseases and simultaneously tracked their movements across a human-impacted landscape as they interacted with human dwellings, fruit plantations, and pig farms. These bats were included as part of a greater study conducting zoonotic viral disease surveillance in over 500 bats of various species at two high-risk human-wildlife interface sites of Myanmar (Valitutto et al. 2020). We aim to identify potential human-wildlife interfaces by evaluating daily movement patterns and spatial distribution of feeding sites for Indian flying fox.
Methods
Study Site
Our study site was the town of Okekan in the Bago region of Myanmar (Lat: 17.499553°, Lon: 95.856097°). Okekan sits in an agricultural landscape dominated by rice growing with small-scale pig farming and various plantations such as rubber, banana, and teak and a mixture of intact and degraded forest in steeper areas. Okekan is one of two large towns in a township with a combined population of approximately 226,000 people. The most common occupation is farming or day laboring on other farms and construction projects. A small number of trees in Okekan are known to support colonies of fruit bats with populations fluctuating greatly across wet and dry seasons. The roost trees chosen for this study sit inside a local police compound surrounded by human dwellings and pig enclosures containing approximately ~ 20 pigs directly underneath the roost trees.
Study Species
The Indian flying fox (Pteropus giganteus) is a large frugivorous bat distributed throughout South Asia from Pakistan to Myanmar (IUCN, 2019). With an average wingspan of 1.3 m and body mass ranging from 0.6 kg to 1.6 kg (Nowak and Walker 1999) it is one of the largest bats in the world and capable of comfortably carrying a GPS transmitter. A common sight in residential and agricultural areas close to water, this bat species is sympatric with humans throughout its range. Even though Pteropus species tend to form large communal roosts, they largely forage individually and have been known to travel long distances of up to 3011 km (Breed et al. 2010; Tidemann and Nelson 2004). P. giganteus is considered an important reservoir for a range of zoonotic diseases and viruses with zoonotic potential including those in the Paramyxovirus and Coronavirus families (Anthony et al. 2017; Epstein et al. 2008). Prior research has indicated that P. giganteus prefer roosting in fragmented forest environments such as those found within village communities, increasing the risk of disease overlap (Hahn et al. 2014). Bats were humanely trapped, handled and sampled according to protocols approved by the Institutional Animal Care and Use Committee of the University of California at Davis (Protocol 19,300) and Smithsonian Institution (Protocol 16–05), and with approvals of Myanmar’s Ministry of Agriculture, Livestock and Irrigation (MAOLI) and Ministry of Natural Resources and Environmental Conservation (MONREC).
The animals were captured over a period of three nights in late April 2018 with a size No. 11 nylon mist net extending about 25 m at one border of a known roost site by trained field personnel in collaboration with MAOLI and MONREC in partnership with the Smithsonian Conservation Biology Institute field team. Nets were monitored continuously after placing at an elevation of approx. 25 m using bamboo poles and bats were removed within ten minutes of capture.
A total of 19 bats were sampled during the first two days of capture for collaring. An additional 10 bats were captured and sampled using the same methods over the following months for viral analysis alone. Samples were taken within 45 min of capture of all animals. Sex, age, morphometrics, reproductive condition, and body condition score were recorded for each animal and a brief physical examination was performed to confirm the lack of external indicators of disease. Flying foxes were classified as either juvenile or adult based on secondary sexual characteristics including presence of penile barbs in the case of males and elongated and white-tipped teats indicating lactation in the case of females. All animals received 10–20 mL of fruit juice solution for hydration before release. The handling time for each individual animal was less than five minutes, including collar placement on selected individuals.
GPS Tracking
Ten apparently healthy adult male bats as determined by physical examination that weighed between 630 and 770 g (mean 697 g) were equipped with GPS tracking collars (LiteTrack 20, SirTrack, New Zealand) weighing 8 g (< 3% of body mass). The collars had an expected accuracy of < 10 m (Forin-Wiart et al. 2015) and were programmed to acquire a GPS fix every 1.5 h throughout the day and night. Data were separated into day and night fixes based on the hours of first and last light. Analyses of foraging movements, described below, were conducted using night time fixes only. While the majority of foraging is nocturnal and the bats mostly roost at night, it is possible that a small number of diurnal foraging sites were excluded from the data or that some nocturnal roosting behavior was considered to be foraging. To further exclude roosting behavior from foraging analysis, an exclusion zone was estimated as the extent of the urban environment around the central roost, extending to cover all dwellings as assessed visually from satellite imagery (Google Earth). This included actual roosting and local flights around the roost site during early morning and late evening. The resulting radius was approximately 700 m around the main roost tree.
The collars were constructed from modified zip-ties with a weak link made from 4 to 0 polydioxanone absorbable suture which was anticipated to decay over 3–6 months based on manufacturer guidelines on internal hydrolysis absorption of suture, which would allow the collar to drop off (Dunn and Phillips 2010). For collar placement, the zip-tie portion of the collar was slid beneath the neck and fastened until two fingers could be placed between the collar and the skin of the neck. Collars were adjusted such that the radio antennae extended down along the back between the shoulders to not interfere with wing movement, and the battery hung ventrally around the neck.
A base station was established in a house approximately 50 m away from the roost tree with direct line of sight to the bats. A VHF antenna was connected to the base station (PinPoint Commander, Lotek Wireless, Havelock North, New Zealand) for remote data download. The base station was set to download data from the collars during the time which the bats were anticipated to be at the roosting tree, from 11:00 to 14:00 every day. The unit was connected to a solar panel and 12-V car battery.
Data were downloaded from the base station once every three weeks. Periodic roost site checks were performed with binoculars to locate as many collared bats as possible; a handheld VHF receiver was also used to identify collared bats present at the site during each visit. Tracking data have been uploaded to an online repository (movebank.org) and are available upon request.
Cluster Analysis
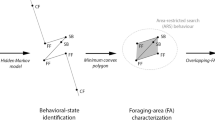
All statistical analysis was carried out in the R programming environment (R Core Development Team 2013). Data were pooled for all individuals. To identify foraging sites and revisitation rates, we used density based spatial clustering methods using the R-packages “dbscan” and “fpc” (Hahsler et al. 2019; Hennig 2015). To define clusters in the data, we set the minimum number of points in a cluster as the log of our sample size and used k-means clustering to determine the most appropriate “reachability distance”, the longest distance between two points that could still be considered to be in the same cluster. This process yielded a minimum number of points = 6 and a reachability distance = 900 m. Points were grouped by cluster membership and plotted on a map of the study site. To further identify areas of intense use (for foraging or roosting) within clusters we generated kernel density utilization distributions using R package “adeHabitatHR” (Calenge 2011), taking the 50% contour as the core are of intense use.
Identifying Foraging Sites
To estimate the number of unique foraging locations visited by each bat we drew a circle of radius 50 m around each GPS location (excluding points at the main roost site), to include the canopy of a large fruit tree and capture small flights around it during foraging. Circles that overlapped were considered to be part of the same location (e.g., a stand of trees close together). Studies on similar sized fruit bats have shown that they typically select a single foraging location for an entire night, making smaller local flights around that area (Markus and Hall 2004; Oleksy et al. 2015; Weber et al. 2015).
Four trained local observers visited three villages that were identified as high-use sites identified from the core area of (50% contour) of utilization distributions. They recorded human population, the species of tree being used, the presence of any roosting bats and the presence of any agricultural animals, crops or human dwellings within 15 m of the focal tree.
Viral and Hematological Sampling
Oral and rectal swabs were collected using sterile polyester-tipped applicators, collected in 500µL viral transport medium (ThermoScientific MicroTest tubes, Fisher Scientific, Pittsburgh, PA, USA) or 500µL TRIzol reagent (Invitrogen TRIzol reagent, Fisher Scientific, Pittsburgh, PA, USA), transported from the field in liquid nitrogen, and transferred to a -800C freezer within five days until time of testing. Hematological samples were obtained with a 1 ml syringe attached to a 25 gage needle. The right cephalic vein was prepped with isopropyl alcohol and approximately 1.5 ml of blood was collected from each individual for additional research sampling.
Viral sample testing was performed at the Myanmar Livestock, Breeding, and the Veterinary Department’s (LBVD) Veterinary Diagnostics Laboratory in Yangon. RNA was extracted using Direct-Zol RNA/DNA extraction kits (Zymo Research Corp., Irvine, CA, USA), and cDNA was prepared using Invitrogen Superscript III (Invitrogen Corp., Fisher Scientific, Pittsburgh, PA, USA). Samples were screened for coronaviruses (CoVs) using two broadly reactive consensus conventional polymerase chain reaction (PCR) assays targeting two non-overlapping fragments (434 bp and 332 bp) of the RNA-dependent RNA polymerase (RdRp) of orfl ab of CoVs (Quan et al. 2010; Watanabe et al. 2010); for filoviruses with a modified degenerate primer set adapted from Zhai, J et al. (2007) targeting the L-gene of the filovirus family for optimized pathogen discovery; for paramyxoviruses utilizing a conventional hemi-nested PCR targeting the polymerase (pol) gene (Tong et al. 2008); and for influenza viruses using two-step PCR using Superscript to make cDNA targeting the M gene (Anthony et al. 2013).
Results
Movement Data
Bats were tracked for an average of 41.5 nights (± 29.81 SD, range: 3 – 83 nights). One collar dropped off, having broken at the weak link, in just over two days and was recovered. The collars collected a total of 2377 location fixes. Using horizontal dilution of precision (HDOP) as a measure of spatial accuracy, we removed locations with HDOP values < 100, leaving a final working data set of 2357 fixes. GPS locations were collected with an average inter-fix interval of 1.77 h (± 0.09 SD), slightly longer than the expected 1.5 h interval. Missing fixes were likely strongly affected by the bats’ position under dense canopy when foraging or roosting. For a summary of GPS collar performance see supplementary material, Table 1. GPS tracking data showed a central-place foraging pattern common across many bat species (Fig. 1). Individuals tagged at a communal roost traveled on average 14.5 km (± 19.71 SD) to feeding sites with an average distance between GPS fixes taken at night of 7.14 km (± 15.65 SD) across all bats. The majority of foraging trips lasted one night only but some individuals took multi-night trips before returning to the roost. One bat traveled 72 km away from the original roost site, returning three days later (Fig. 1). Selection of foraging sites differed across individuals even though they shared the same roost. There were only two cases of bats visiting the same foraging location and only one case of two collared bats being present in the same area on the same night. All foraging locations occurred within human-modified landscapes, often on agricultural land or fruiting trees in residential areas where small-scale pig farming is the main source of income for villagers. Although roosting together at a central location for the majority of the study period, bats traveled to different foraging sites, each showing a preference for particular places and sometimes roosting elsewhere for a number of nights (mean percentage of total tracking days spent on central roost: 81.74% ± 14.6 SD).
Clustering analysis revealed six broad clusters of bat foraging locations (Fig. 2a) within which core areas of usage were identified (Fig. 2b) and selected for site visits. Analysis of individual foraging sites, using 50 m circles around GPS locations, revealed 207 unique locations visited across all bats (Table 1, Fig S1).
Site inspections revealed that bats were targeting red silk cotton trees (Bombax ceiba) for feeding and roosting and mango trees for feeding. Site 1 had the largest human population (~ 3000 people). Pig farming was common at this site with every house containing at least two pigs. At this site two households were located directly under the main foraging tree and a further 10 households within 15 m of the center of the foraging area. During the site inspection, 150 bats were counted in a red silk cotton tree. Rats (species unknown) were also observed in the center of the foraging area. At Site 2, the human population was lower (~ 150 people) and again pig farming was common across all households. Two larger farms in the area had approximately 100 pigs in each. The foraging area was centered on a mango tree inside of a monastery compound containing 5 houses and 6 pig farms located within a 15 m radius. The area is known to be frequented by large flocks of pigeons. Site 3 was a village of 400 households and 2000 people. The center of the foraging area was located within a large farm. No households were present within 15 m of the center, the nearest being 800 m away. There were rice paddy fields within 15 m of the center which are regularly worked by two families. No pig farms or other animals were observed within the foraging area.
Viral Sampling
Of the 29 bats sampled, no positive results were recorded for any of the pathogens for which we tested by PCR.
Discussion
This study aimed to evaluate the movement patterns in a colony of Indian flying foxes with a focus on identification of human-wildlife interfaces in Myanmar. A previous study evaluating the general viral biome of bats across Myanmar revealed a wide host of viruses present across many bat species (He et al. 2013b), and in nearby countries including China and Bangladesh, fruit bat populations have been documented carrying emerging infectious diseases (He et al. 2013b; Hu et al. 2014; Luby et al. 2006). Understanding the potential for disease transmission in bats of this region, we believe determining the natural movement patterns of the species is an essential step in the prediction and prevention of zoonotic disease transmission.
The observed pattern of individuals dispersing across the local landscape to different foraging areas and returning to a shared roost identifies areas of human-wildlife interface for this Pteropus colony during the study period. Observations from site visits by researchers on the ground confirm the close presence of humans, livestock, and rodents to foraging areas; however, further evaluation is required to determine the presence of zoonotic disease and risk of interspecies transmission. Across the study landscape, these 10 bats visited a total of 207 unique locations. Although it is unlikely that every individual bat in the colony traveled alone to forage every night, the individual nature of foraging site selection we observed in our sample suggests that, if extrapolated to the total number of bats at this one roost, this could mean the entirety of the roost could be traveling to hundreds of locations over a short period of time. The movement characteristics of bats in this study such as selecting a single location for foraging each night, occasionally undertaking long distance flight, and the average distances moved between a roost and foraging site, align with findings on other Pteropus species. (Banack and Grant 2002; Roberts et al. 2012; Spencer et al. 1991).
Visits to selected foraging sites by researchers on the ground revealed intense foraging on successive nights in areas closely associated with humans and agriculture, often directly above livestock and dwellings. The density of livestock (roughly to 2 pigs per household), the proximity of houses to bat foraging and roosting trees, and the presence of large numbers of rodents create a perfect interface site between bats, livestock, and humans. Even in locations such as Site 3 where livestock and dwellings were further removed from the foraging tree, humans still worked in rice fields directly underneath the foraging trees. In addition, the direct consumption of bats as a food source is not uncommon in this area and the sale and consumption of wildlife products is widespread throughout Myanmar (McEvoy et al. 2019).
With a stable roost at a particular site, locals may assume that bats in this area are the same individuals, but our tracking data suggest a more dynamic situation. The presence of secondary roosts in the landscape was indicated by the finding that bats spent the majority of nights on the central roost (81.74% ± 14.6 SD) but not all nights. For example, at Site 1, 150 bats were observed roosting during the day. This site was within 10 km of our main roost site and not only a foraging site but also contained a secondary roost. Movement data revealed two collared bats moving between the two, demonstrating flow of individuals between roosts. Although an extensive survey of all potential roost sites within the local landscape was beyond the scope of this study, the suggested existence of multiple communal roost sites in the landscape where roost membership appears to be fluid, aligns with results from studies on other Pteropus species (Roberts et al. 2012; Spencer et al. 1991).
Our movement data were limited by a number of factors. Although P. giganteus is one of the largest species of bats in the world, we are still limited by the size of the transmitter that can be attached. For the purposes of this study, we chose to collect high-accuracy, 1.5 hourly GPS data to identify specific foraging sites. This limited our data collection period to a maximum of three months. In future studies, a mixture of sampling rates could be used and by sacrificing data frequency we could potentially track individuals throughout an entire year. In addition, the collar design required modification to allow drop-off of the collar without recapturing individual animals which would be logistically challenging and stressful to the animals. This modification was performed relying on the assumption that degradation of the suture material would occur; however, the published information on dissolvable sutures relies on hydrolysis of material within body tissues, while this study relied on environmental conditions to degrade suture material. While one bat removed a collar within 24 h of placement, all other collars remained in place for the duration of the study and based on movement data it was presumed that animals remained generally uninhibited to pursue feeding opportunities by the presence of the collars for the 3 months of data collection. However, it is unknown if the collars caused a reduction in normal movement activity nor if the collars caused any physical damage to the bats. Future visits to the site failed to identify collared bats in the colony.
The base station/VHF download system for collecting data was necessary due to the small size of the transmitters. More battery power, and hence more weight, would have been required to transmit to a satellite. This meant that if a bat did not return to the roost where the base station was located, whatever data it was carrying were lost and its fate could not be determined. A wider array of base stations at different potential roost sites or a system that transmits data via GSM networks would improve our ability to capture more complex movement patterns.
The size and the position of the collars on this species proved to be somewhat challenging. The location of the GPS unit on the collar was toward the right shoulder of many bats, and in some cases it ended up ventrally located due to the small neck circumference of individuals. This limited the accuracy of the GPS signals during flight. Accurate measures of neck circumference with pre-project capture attempts, while risking disruption of the roost, may provide more accurate collar design in the future and improve signal accuracy.
In a study to identify broader coronaviral diversity of bats, Anthony et. al. (2017) recommended sampling greater than 400 individuals to optimize the full diversity of coronaviruses present within any particular species. The bats examined in this study were part of a larger study conducting zoonotic viral disease surveillance to determine prevalence and potential for cross-species transmission (Valitutto et al. 2020). Multiple bat species were included with samples collected from over 400 individual bats at two high-risk interface sites in Myanmar. Per the results of that study, none of the viruses evaluated were detected in the bats of this GPS study. However, novel and known coronaviruses and paramyxoviruses were detected in other bat species (Scotophilus sp. Taphozous sp., Chaerephon sp., Hipposideros sp) within the same region as the GPS bat colony (Valitutto et al, 2020, Valitutto et al., in review). Regardless of the results of the broader study and the large multi-species populations sampled, to fully appreciate the potential for this bat colony to host a zoonotic virus and the risk of transmission, further independent studies are needed with a primary focus on this population of Indian flying fox. The physiologic and geographic diversity of bats is profound, and thus, each species has a different potential for both hosting and spreading zoonotic diseases (Calisher et al. 2006). To understand the risk of zoonotic disease transmission, individual bat species and populations must be examined for their movement patterns, foraging behaviors, colony structure, and dynamics of hosting a zoonotic virus (e.g., seasonal variation, intra-species transmission, gender, health status, biological secretions, etc.) (Calisher et al, 2006). Given the interspecies differences, the lack of viral detection may be attributed to the small sample size but other reasons cannot be ruled out including collection and processing errors, seasonal variation, capture bias (behavioral difference of sick and/or virus-positive bats), or that this is a genuinely negative colony.
Our movement data identified individuality in foraging site selection, and considering the distances flown each night, there are potentially many more human-interface sites along the daily and seasonal movement paths of these bats. Expanding the study to include more roosts would allow us to determine the flow of individuals between roost sites. All of the bats tracked in this study were deemed healthy on physical examination with no viruses detected of the four viral families screened. Tracking a greater number of bats with different disease statuses would also help to evaluate if the identified patterns of movement behavior and site selection are identical in bats that are carrying pathogens (with or without symptoms) or if transmission-relevant movement behavior shifts while carrying infectious pathogens, as seen in other species (Hawley et al. 2007; Stockmaier et al. 2021; Wilcox et al. 2014).
Conclusions
Our results highlight the importance of understanding the movement behavior of Pteropus giganteus in order to identify human-interface sites and assess their potential for the spread of zoonotic disease to livestock and humans. GPS tracking allowed us to determine the flow of individuals from roosts to foraging sites and to assess the conditions on the ground at these sites. The lack of detection of any screened viruses in this bat colony highlights the importance of disease surveillance programs to be mindful of the difference between bat species in study design and focus. Public health programs should target areas with sizeable bat roosts and foraging areas for education, outreach, and vaccination. Further work is needed to fully understand the risk of disease transmission between bats within a colony as well as between bats and humans and livestock at these interface sites. Natural behavioral studies would help determine if there are migratory and resident sub-populations of this bat colony and if bats from this region intermix with those in areas known to have had outbreaks of infectious diseases. Frugivorous bats are of huge ecological importance in tropical ecosystems and their populations are under threat from many directions (Allen-Wardell et al. 1998; Corlett 2009). By more fully understanding the links between their movement behavior and the potential for interface with humans, we can protect human health while ensuring that bats can continue to thrive in these landscapes.
References
Abedi-Lartey M, Dechmann DK, Wikelski M, Scharf AK, Fahr J (2016) Long-distance seed dispersal by straw-coloured fruit bats varies by season and landscape. Global Ecology and Conservation 7:12–24
Allen-Wardell, G., Bernhardt, P., Bitner, R., Burquez, A., Buchmann, S., Cane, J., Cox, P.A., Dalton, V., Feinsinger, P., Ingram, M., 1998. The potential consequences of pollinator declines on the conservation of biodiversity and stability of food crop yields. Conservation Biology, 8–17.
Anthony, S.J., Epstein, J.H., Murray, K.A., Navarrete-Macias, I., Zambrana-Torrelio, C.M., Solovyov, A., Ojeda-Flores, R., Arrigo, N.C., Islam, A., Khan, S.A., 2013. A strategy to estimate unknown viral diversity in mammals. MBio 4.
Anthony, S.J., Johnson, C.K., Greig, D.J., Kramer, S., Che, X., Wells, H., Hicks, A.L., Joly, D.O., Wolfe, N.D., Daszak, P., 2017. Global patterns in coronavirus diversity. Virus evolution 3.
Aziz SA, McConkey KR, Tanalgo K, Sritongchuay T, Low M-R, Yong JY, Mildenstein TL, Nuevo-Diego CE, Lim V-C, Racey PA (2021) The critical importance of Old World fruit bats for healthy ecosystems and economies. Frontiers in Ecology and Evolution 9:181
Banack SA, Grant GS (2002) Spatial and Temporal Movement Patterns of the Flying Fox, Pteropus tonganus, in American Samoa. The Journal of Wildlife Management 66:1154–1163
Bates, P.J., Nwe, T., Pearch, M.J., Swe, K.M., Bu, S.H., Tun, T., 2000. A review of bat research in Myanmar (Burma) and results of a recent survey. Acta Chiropterologica 1.
Bhagwat T, Hess A, Horning N, Khaing T, Thein ZM, Aung KM, Aung KH, Phyo P, Tun YL, Oo AH, Neil A, Thu WM, Songer M, Connette KL, Bernd A, Huang Q, Connette G, Leimgruber P (2017) Losing a jewel-Rapid declines in Myanmar’s intact forests from 2002–2014. Plos One 12:22
Breed AC, Field HE, Smith CS, Edmonston J, Meers J (2010) Bats without borders: long-distance movements and implications for disease risk management. EcoHealth 7:204–212
Brierley L, Vonhof MJ, Olival KJ, Daszak P, Jones KE (2016) Quantifying global drivers of zoonotic bat viruses: a process-based perspective. The American Naturalist 187:E53–E64
Calderón-Capote, M.C., Dechmann, D.K., Fahr, J., Wikelski, M., Kays, R., O'Mara, M.T., 2020. Foraging movements are density-independent among straw-coloured fruit bats. Royal Society Open Science 7, 200274.
Calenge, C., 2011. Home range estimation in R: the adehabitatHR package. Office national de la classe et de la faune sauvage: Saint Benoist, Auffargis, France.
Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T (2006) Bats: important reservoir hosts of emerging viruses. Clinical Microbiology Reviews 19:531–545
Chong, H.T., Abdullah, S., Tan, C.T., 2009. Nipah virus and bats. Neurology Asia 14.
Chua KB (2003) Nipah virus outbreak in Malaysia. Journal of Clinical Virology 26:265–275
Corlett RT (2009) Seed Dispersal Distances and Plant Migration Potential in Tropical East Asia. Biotropica 41:592–598
Daversa, D.R., Fenton, A., Dell, A.I., Garner, T.W.J., Manica, A., 2017. Infections on the move: how transient phases of host movement influence disease spread. Proceedings of the Royal Society B: Biological Sciences 284.
Dunn, D.L., Phillips, J., 2010. Wound Closure Manual. University of Minnesota, 6–13.
Epstein, J., Rahman, S., Pulliam, J., Hassan, S., Halpin, K., Smith, C., Jamaluddin, A., Chua, K., Field, H., Hyatt, A., 2008. The emergence of Nipah virus in Malaysia: the role of pteropus bats as hosts and agricultural expansion as a key factor for zoonotic spillover. International Journal of Infectious Diseases 12, e46.
Epstein JH, Olival KJ, Pulliam JR, Smith C, Westrum J, Hughes T, Dobson AP, Zubaid A, Rahman SA, Basir MM (2009) Pteropus vampyrus, a hunted migratory species with a multinational home-range and a need for regional management. Journal of Applied Ecology 46:991–1002
Fahr, J., Abedi-Lartey, M., Esch, T., Machwitz, M., Suu-Ire, R., Wikelski, M., Dechmann, D.K., 2015. Pronounced seasonal changes in the movement ecology of a highly gregarious central-place forager, the African straw-coloured fruit bat (Eidolon helvum). Plos One 10, e0138985.
Feare CJ (2010) Role of Wild Birds in the Spread of Highly Pathogenic Avian Influenza Virus H5N1 and Implications for Global Surveillance. Avian Diseases 54:201–212
Forin-Wiart, M.-A., Hubert, P., Sirguey, P., Poulle, M.-L., 2015. Performance and accuracy of lightweight and low-cost GPS data loggers according to antenna positions, fix intervals, habitats and animal movements. Plos One 10, e0129271.
Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y, Shi Z (2012) Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. Journal of Virology 86:4620–4630
Giles JR, Eby P, Parry H, Peel AJ, Plowright RK, Westcott DA, McCallum H (2018) Environmental drivers of spatiotemporal foraging intensity in fruit bats and implications for Hendra virus ecology. Scientific Reports 8:9555
Gumal MT (2004) Diurnal home range and roosting trees of a maternity colony of Pteropus vampyrus natunae (Chiroptera: Pteropodidae) in Sedilu, Sarawak. Journal of Tropical Ecology 20:247–258
Hahn MB, Epstein JH, Gurley ES, Islam MS, Luby SP, Daszak P, Patz JA (2014) Roosting behaviour and habitat selection of Pteropus giganteus reveal potential links to Nipah virus epidemiology. Journal of Applied Ecology 51:376–387
Hahsler, M., Piekenbrock, M., Arya, S., Mount, D., 2019. Density-based clustering of applications with noise (DBSCAN) and related algorithms. CRAN.
Harten L, Katz A, Goldshtein A, Handel M, Yovel Y (2020) The ontogeny of a mammalian cognitive map in the real world. Science 369:194–197
Hasebe F, Le TQM (2007) Surveillance of bats as reservoir hosts of emerging zoonotic viruses in Vietnam. Tropical Medicine and Health 35:51–53
Hawley D, Davis A, Dhondt A (2007) Transmission-relevant behaviours shift with pathogen infection in wild house finches (Carpodacus mexicanus). Canadian Journal of Zoology 85:752–757
He B, Fan Q, Yang F, Hu T, Qiu W, Feng Y, Li Z, Li Y, Zhang F, Guo H (2013a) Hepatitis virus in long-fingered bats. Myanmar. Emerging Infectious Diseases 19:638
He, B., Li, Z., Yang, F., Zheng, J., Feng, Y., Guo, H., Li, Y., Wang, Y., Su, N., Zhang, F., 2013b. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. Plos One 8, e61950.
Hennig, C., 2015. Package ‘fpc’. Available at: Available at: https://cran.r-project.org/web/packages/fpc/index.htmlENT91.
Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, Niezgoda M, Rupprecht C, Bresee J, Breiman RF (2004) Nipah virus encephalitis reemergence, Bangladesh. Emerging Infectious Diseases 10:2082
Hu T, Qiu W, He B, Zhang Y, Yu J, Liang X, Zhang W, Chen G, Zhang Y, Wang Y (2014) Characterization of a novel orthoreovirus isolated from fruit bat, China. BMC Microbiology 14:293
Hughes JM, Wilson ME, Halpin K, Hyatt AD, Plowright RK, Epstein JH, Daszak P, Field HE, Wang L, Daniels PW (2007) Emerging viruses: coming in on a wrinkled wing and a prayer. Clinical Infectious Diseases 44:711–717
Hutson, A., Mickleburgh, S., Racey, P., 2001. Microchiropteran bats: global status survey and conservation action plan. IUCN/SSC Chiroptera Specialist Group, Gland, Switzerland and Cambridge, UK.
Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P (2008) Global trends in emerging infectious diseases. Nature 451:990
Kruse H, Kirkemo A-M, Handeland K (2004) Wildlife as source of zoonotic infections. Emerging Infectious Diseases 10:2067
Luby SP, Rahman M, Hossain MJ, Blum LS, Husain MM, Gurley E, Khan R, Ahmed B-N, Rahman S, Nahar N (2006) Foodborne transmission of Nipah virus, Bangladesh. Emerging Infectious Diseases 12:1888
Machhi, J., Herskovitz, J., Senan, A.M., Dutta, D., Nath, B., Oleynikov, M.D., Blomberg, W.R., Meigs, D.D., Hasan, M., Patel, M., Kline, P., Chang, R.C.-C., Chang, L., Gendelman, H.E., Kevadiya, B.D., 2020. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. Journal of Neuroimmune Pharmacology.
Mackenzie J, Field H, Guyatt K (2003) Managing emerging diseases borne by fruit bats (flying foxes), with particular reference to henipaviruses and Australian bat lyssavirus. Journal of Applied Microbiology 94:59S-69S
Mackenzie J, Williams D (2009) The zoonotic flaviviruses of Southern, South-Eastern and Eastern Asia, and australasia: the potential for emergent viruses. Zoonoses and Public Health 56:338–356
Markus N, Hall L (2004) Foraging behaviour of the black flying-fox (<emph type="2">Pteropus alecto</emph>) in the urban landscape of Brisbane, Queensland. Wildlife Research 31:345–355
McEvoy, J.F., Connette, G., Huang, Q., Soe, P., Pyone, K.H.H., Valitutto, M., Htun, Y.L., Lin, A.N., Thant, A.L., Htun, W.Y., Paing, K.H., Swe, K.K., Aung, M., Min, S., Songer, M., Leimgruber, P., 2019. Two sides of the same coin – Wildmeat consumption and illegal wildlife trade at the crossroads of Asia. Biological Conservation 238, 108197.
McMichael L, Edson D, Mayer D, McLaughlin A, Goldspink L, Vidgen ME, Kopp S, Meers J, Field H (2016) Temporal variation in physiological biomarkers in Black flying-foxes (Pteropus alecto), Australia. EcoHealth 13:49–59
Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, AlHakeem R, Durosinloun A, Al Asmari M, Islam A (2013) Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerging Infectious Diseases 19:1819
Mickleburgh SP, Hutson AM, Racey PA (1992) Old World fruit bats - An action plan for their conservation. Gland, Switzerland: IUCN
Mickleburgh SP, Hutson AM, Racey PA (2002) A review of the global conservation status of bats. Oryx 36:18–34
Nijman V (2010) An overview of international wildlife trade from Southeast Asia. Biodiversity and Conservation 19:1101–1114
Nijman V, Shepherd CR (2014) Emergence of Mong La on the Myanmar-China border as a global hub for the international trade in ivory and elephant parts. Biological Conservation 179:17–22
Nowak, R.M., Walker, E.P., 1999. Walker's Mammals of the World. JHU press.
Oleksy, R., Giuggioli, L., McKetterick, T.J., Racey, P.A., Jones, G., 2017. Flying foxes create extensive seed shadows and enhance germination success of pioneer plant species in deforested Madagascan landscapes. Plos One 12, e0184023.
Oleksy R, Racey PA, Jones G (2015) High-resolution GPS tracking reveals habitat selection and the potential for long-distance seed dispersal by Madagascan flying foxes Pteropus rufus. Global Ecology and Conservation 3:678–692
Olival KJ, Islam A, Yu M, Anthony SJ, Epstein JH, Khan SA, Khan SU, Crameri G, Wang L-F, Lipkin WI, Luby SP, Daszak P (2013) Ebola virus antibodies in fruit bats, bangladesh. Emerging Infectious Diseases 19:270–273
Olival, K.J., Weekley, C.C., Daszak, P., 2015. Are bats really ‘special’as viral reservoirs? What we know and need to know. Bats and viruses, 281–294.
Oo KS, Win HL, Bates PJ, Pearch M (2017) The ecology and distribution of Flying Foxes (Chiroptera: Pteropodidae: Pteropus) in Tanintharyi Region, Myanmar with a first mainland record of Pteropus hypomelanus geminorum from Myeik. Journal of Threatened Taxa 9:10528–10537
Plowright RK, Foley P, Field HE, Dobson AP, Foley JE, Eby P, Daszak P (2011) Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (<i>Pteropus</i> spp.). Proceedings of the Royal Society B: Biological Sciences 278:3703–3712
Quan, P.-L., Firth, C., Street, C., Henriquez, J.A., Petrosov, A., Tashmukhamedova, A., Hutchison, S.K., Egholm, M., Osinubi, M.O., Niezgoda, M., 2010. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio 1.
R Core Development Team, 2013. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Roberts BJ, Catterall CP, Eby P, Kanowski J (2012) Long-distance and frequent movements of the flying-fox Pteropus poliocephalus: implications for management. Plos One 7:e42532–e42532
Sapir N, Horvitz N, Dechmann DK, Fahr J, Wikelski M (2014) Commuting fruit bats beneficially modulate their flight in relation to wind. Proceedings of the Royal Society B: Biological Sciences 281:20140018
Schuh AJ, Amman BR, Jones ME, Sealy TK, Uebelhoer LS, Spengler JR, Martin BE, Coleman-McCray JAD, Nichol ST, Towner JS (2017) Modelling filovirus maintenance in nature by experimental transmission of Marburg virus between Egyptian rousette bats. Nature Communications 8:14446
Smith, I., Broos, A., de Jong, C., Zeddeman, A., Smith, C., Smith, G., Moore, F., Barr, J., Crameri, G., Marsh, G., 2011. Identifying Hendra virus diversity in pteropid bats. Plos One 6, e25275.
Spencer, H., Palmer, C., Parry-Jones, K., 1991. Movements of Fruit-bats in eastern Australia, determined by using radio-tracking. Wildlife Research - WILDLIFE RES 18.
Stockmaier, S., Stroeymeyt, N., Shattuck, E.C., Hawley, D.M., Meyers, L.A., Bolnick, D.I., 2021. Infectious diseases and social distancing in nature. Science 371, eabc8881.
Struebig MJ, Rossiter SJ, Bates PJ, Kingston T, Lin Oo SS, Nwe AA, Aung MM, Win SS, Mya KM (2005) Results of a recent bat survey in Upper Myanmar including new records from the Kachin forests. Acta Chiropterologica 7:147–163
Syafiq M, Atiqah ARN, Ghazali A, Asmah S, Yahya MS, Aziz N, Puan CL, Azhar B (2016) Responses of tropical fruit bats to monoculture and polyculture farming in oil palm smallholdings. Acta Oecologica-International Journal of Ecology 74:11–18
Tidemann CR, Nelson JE (2004) Long-distance movements of the grey-headed flying fox (Pteropus poliocephalus). Journal of Zoology 263:141–146
Toledo S, Shohami D, Schiffner I, Lourie E, Orchan Y, Bartan Y, Nathan R (2020) Cognitive map–based navigation in wild bats revealed by a new high-throughput tracking system. Science 369:188–193
Tong S, Chern S-WW, Li Y, Pallansch MA, Anderson LJ (2008) Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. Journal of Clinical Microbiology 46:2652–2658
Valitutto, M.T., Aung, O., Tun, K.Y.N., Vodzak, M.E., Zimmerman, D., Yu, J.H., Win, Y.T., Maw, M.T., Thein, W.Z., Win, H.H., Dhanota, J., Ontiveros, V., Smith, B., Tremeau-Brevard, A., Goldstein, T., Johnson, C.K., Murray, S., Mazet, J., 2020. Detection of novel coronaviruses in bats in Myanmar. Plos One 15, e0230802.
van Toor ML, O’Mara MT, Abedi-Lartey M, Wikelski M, Fahr J, Dechmann DK (2019) Linking colony size with quantitative estimates of ecosystem services of African fruit bats. Current Biology 29:R237–R238
Watanabe S, Masangkay JS, Nagata N, Morikawa S, Mizutani T, Fukushi S, Alviola P, Omatsu T, Ueda N, Iha K (2010) Bat coronaviruses and experimental infection of bats, the Philippines. Emerging Infectious Diseases 16:1217
Weber N, Duengkae P, Fahr J, Dechmann DK, Phengsakul P, Khumbucha W, Siriaroonrat B, Wacharapluesadee S, Maneeorn P, Wikelski M (2015) High-resolution GPS tracking of Lyle’s flying fox between temples and orchards in central Thailand. The Journal of Wildlife Management 79:957–968
Welbergen JA, Meade J, Field HE, Edson D, McMichael L, Shoo LP, Praszczalek J, Smith C, Martin JM (2020) Extreme mobility of the world’s largest flying mammals creates key challenges for management and conservation. BMC Biology 18:1–13
Wilcox A, Warnecke L, Turner JM, McGuire LP, Jameson JW, Misra V, Bollinger TC, Willis CKR (2014) Behaviour of hibernating little brown bats experimentally inoculated with the pathogen that causes white-nose syndrome. Animal Behaviour 88:157–164
Willoughby, A., Phelps, K., Olival, K., Consortium, P., 2017. A comparative analysis of viral richness and viral sharing in cave-roosting bats. Diversity 9, 35.
Young, C.C., Olival, K.J., 2016. Optimizing viral discovery in bats. Plos One 11, e0149237.
Zhai J, Palacios G, Towner JS, Jabado O, Kapoor V, Venter M, Grolla A, Briese T, Paweska J, Swanepoel R (2007) Rapid molecular strategy for filovirus detection and characterization. Journal of Clinical Microbiology 45:224–226
Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273
Acknowledgements
The authors wish to acknowledge the Smithsonian Women’s Committee and Judy and John W. McCarter, Jr. for their vital contributions supporting the project development and completion. This study was made possible by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (cooperative agreement number AID-OAA-A-14-00102). The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government. Additional support for manuscript preparation was provided by the Smithsonian Institution, the Smithsonian Movement of Life Initiative, and the Morris Animal Foundation and Dennis and Connie Keller through a training partnership. The support provided by the Myanmar field team in bat restraint, surveillance, and sample processing was vital for project completion and the authors wish to thank them for their significant contributions to this work. The authors declare no conflicts of interest in the publication of this work.
Author information
Authors and Affiliations
Contributions
J.C.K., J.F.M., and M.T.V. were responsible for project design and data analysis, as well as manuscript drafting and completion. K.Y.N.T., Y.T.W., M.T.M., W.Z.T., and H.H.W. were primarily responsible for data collection in Myanmar and primary viral data analysis. O.A., K.Y.N.T. and A.M.C. coordinated project development, location sourcing, and staff support in Myanmar as well as manuscript review. M.E.V. and S.M. assisted with manuscript formation and project oversight.
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
McEvoy, J.F., Kishbaugh, J.C., Valitutto, M.T. et al. Movements of Indian Flying Fox in Myanmar as a Guide to Human-Bat Interface Sites. EcoHealth 18, 204–216 (2021). https://doi.org/10.1007/s10393-021-01544-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10393-021-01544-w