Abstract
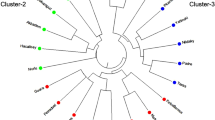
Genetic diversity is an important measure for the improvement of many crop species including Brassica. This study evaluated the genetic divergence among six Brassica species using simple sequence repeats (SSR). Ten SSR markers produced overall 21 alleles with an average of 2.1 alleles per primer. Out of 21, 18 alleles showed polymorphism (85.71%) and 3 alleles showed monomorphism (14.28%). A similarity matrix was constructed using Popgen32 software. Genetic identity ranged from 33.33 to 76.19%. B. nigra and B. campestris showed the highest identity, while the lowest identity was observed between B. campestris and B. oleracea. Sizes of amplified alleles were ranged from 70 to 290 bp. Polymorphic information content (PIC) varied from 0.37 to 0.71, with an average of 0.66 per primer. A dendrogram classified the genotypes into two main clusters. Cluster-A is further divided into cluster-C, which consists of B. carinata and B. oleracea. B. napus and B. juncea each form an independent cluster. Cluster-B consists of B. nigra and B. campestris, meaning that these two species are closely related to each other. The results indicated that these species can be isolated from each other at the molecular level by using molecular markers.
Zusammenfassung
Die genetische Diversität ist ein wichtiges Maß für die Verbesserung vieler Kulturpflanzen einschließlich Brassica. In der vorliegenden Studie wurde die genetische Divergenz zwischen 6 Brassica-Arten unter der Verwendung von Mikrosatelliten („simple sequence repeats“, SSR) untersucht. Dabei produzierten 10 SSR-Marker insgesamt 21 Allele mit einem Durchschnitt von 2,1 Allelen pro Primer. Von den 21 Allelen zeigten 18 Polymorphismus (85,71 %), und 3 Allele zeigten Monomorphismus (14,28 %). Mit der Popgen32-Software wurde eine Ähnlichkeitsmatrix erstellt. Die genetische Identität lag im Bereich von 33,33–76,19 %. Die höchste Identität wiesen B. nigra und B. campestris auf, während die niedrigste Identität zwischen B. campestris und B. oleracea beobachtet wurde. Die Größen der amplifizierten Allele lagen im Bereich von 70–290 bp. Der polymorphe Informationsgehalt (PIC) variierte von 0,37–0,71, mit einem Durchschnitt von 0,66 pro Primer. Ein Dendrogramm klassifizierte die Genotypen in 2 Hauptcluster. Cluster-A wird weiter unterteilt in Cluster-C, der aus B. carinata und B. oleracea besteht. Jeweils einen unabhängigen Cluster bilden B. napus und B. juncea. Cluster-B besteht aus B. nigra und B. campestris, was bedeutet, dass diese beiden Arten eng miteinander verwandt sind. Die Ergebnisse zeigten, dass diese Spezies auf molekularer Ebene unter Verwendung molekularer Marker voneinander isoliert werden können.

Similar content being viewed by others

References
Abbas SJ, Farhatullah MK, Khan I, Munir I (2009) Molecular analysis of genetic diversity in Brassica species. Pak J Bot 41:167–176
Agrama H, Tuinstra M (2003) Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. Afr J Biotechnol 2:334–340
Ayana A, Bryngelsson T, Bekele E (2000) Genetic variation of Ethiopian and Eritrean sorghum (Sorghum bicolor (L.) Moench) germplasm assessed by random amplified polymorphic DNA (RAPD). Genet Resour Crop Evol 47:471–482
Birmeta G, Nybom H, Bekele E (2002) RAPD analysis of genetic diversity among clones of the Ethiopian crop plant Ensete ventricosum. Euphytica 124:315–325
Cartea ME, Picoaga A, Soengas P, Ordás A (2003) Morphological characterization of kale populations from northwestern Spain. Euphytica 129:25–32
Darvishzadeh R, Azizi M, Hatami-Maleki H, Bernousi I, Mandoulakani BA, Jafari M, Sarrafi A (2010) Molecular characterization and similarity relationships among sunflower (Helianthus annuus L.) inbred lines using some mapped simple sequence repeats. Afr J Biotechnol 9:7280–7288
Edwards D, Forster JW, Chagné D, Batley J (2007) What Are SNPs? Assoc Mapp Plants. https://doi.org/10.1007/978-0-387-36011-9_3
Harbaum B, Hubbermann EM, Wolff C, Herges R, Zhu Z, Schwarz K (2007) Identification of Flavonoids and Hydroxycinnamic acids in Pak Choi varieties (brassica campestris L. ssp. chinensis var. communis) by HPLC–ESI-MS n and NMR and their Quantification by HPLC–DAD. J Agric Food Chem 55:8251–8260
Hoshino AA, Bravo JP, Morelli KA, Nobile PM (2012) Microsatellites as tools for genetic diversity analysis. In: Caliskan M (ed) Genetic diversity in microorganisms. Intech Open, London
Hvarleva T, Bakalova A, Chepinski I, Hristova-Cherbadji M, Hristov M, Atanasov A (2007) Characterization of Bulgarian sunflower cultivars and inbred lines with microsatellite markers. Biotechnol Biotechnol Equip 21:408–412
Iqbal S, Hamim I, Haque S, Nath UK (2015) Genetic diversity analysis of mustard (Brassica spp.) germplasm using molecular marker for selection of short duration genotypes. Afr J Biotechnol 14:1439–1448
Kanwal M, Nawaz I (2014) Genetic diversity analysis of Brassica napus/Brassica campestris progenies using microsatellite markers. Pak J Bot 46:779–787
Kong L, Dong J, Hart G (2000) Characteristics, linkage-map positions, and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs). Theor Appl Genet 101:438–448
Liu K, Muse SV (2005) PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mailer R, Scarth R, Fristensky B (1994) Discrimination among cultivars of rapeseed (Brassica napus L.) using DNA polymorphisms amplified from arbitrary primers. Theor Appl Genet 87:697–704
Morin PA, Luikart G, Wayne RK (2004) SNPs in ecology, evolution and conservation. Trends Ecol Evol (Amst) 19(4):208–216. https://doi.org/10.1016/j.tree.2004.01.009
Murray SC, Rooney WL, Hamblin MT, Mitchell SE, Kresovich S (2009) Sweet sorghum genetic diversity and association mapping for brix and height. Plant Genome 2:48–62
Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theor Appl Genet 97:1248–1255
Plieske J, Struss D (2001) Microsatellite markers for genome analysis in Brassica. I. Development in Brassica napus and abundance in Brassicaceae species. Theor Appl Genet 102:689–694
Pradhan A, Yan G, Plummer J (2004) Development of DNA fingerprinting keys for the identification of radish cultivars. Aust J Exp Agric 44:95–102
Rabbani MA, Iwabuchi A, Murakami Y, Suzuki T, Takayanagi K (1998) Genetic diversity in mustard (Brassica juncea L.) germplasm from Pakistan as determined by RAPDs. Euphytica 103:235–242
Ramos-Gómez S, Busto MD, Perez-Mateos M, Ortega N (2014) Development of a method to recovery and amplification DNA by real-time PCR from commercial vegetable oils. Food Chem 158:374–383
Raza A, Shaukat H, Ali Q, Habib M (2018) Assessment of RAPD markers to analyse the genetic diversity among sunflower (Helianthus annuus L.) genotypes. Turk J Agric Food Sci Technol 6:107–111
Ren J, McFerson JR, Li R, Kresovich S, Lamboy WF (1995) Identities and relationships among Chinese vegetable Brassicas as determined by random amplified polymorphic DNA markers. J Am Soc Hortic Sci 120:548–555
Riaz A, Li G, Quresh Z, Swati M, Quiros C (2001) Genetic diversity of oilseed Brassica napus inbred lines based on sequence-related amplified polymorphism and its relation to hybrid performance. Plant Breed 120:411–415
Shengwu H, Ovesna J, Kucera L, Kucera V, Vyvadilová M (2003) Evaluation of genetic diversity of Brassica napus germplasm from China and Europe assessed by RAPD markers. Plant Soil Environ 49:106–113
Sobotka R, Dolanska L, Curn V, Ovesná J (2004) Fluorescence-based AFLPs occur as the most suitable marker system for oilseed rape cultivar identification. J Appl Genet 45:161–174
Tang S, Knapp SJ (2003) Microsatellites uncover extraordinary diversity in native American land races and wild populations of cultivated sunflower. Theor Appl Genet 106:990–1003
Teklewold A, Becker HC (2006) Geographic pattern of genetic diversity among 43 Ethiopian mustard (Brassica carinata A. Braun) accessions as revealed by RAPD analysis. Genet Resour Crop Evol 53(6):1173–1185. https://doi.org/10.1007/s10722-005-2011-4
Tommasini L et al (2003) The development of multiplex simple sequence repeat (SSR) markers to complement distinctness, uniformity and stability testing of rape (Brassica napus L.) varieties. Theor Appl Genet 106:1091–1101
Tonguç M, Griffiths PD (2004) Genetic relationships of Brassica vegetables determined using database derived simple sequence repeats. Euphytica 137:193–201
Tsunoda S, Hinata K, Gómez-Campo C (1980) Brassica crops and wild allies. Biology and breeding. Bull Torrey Bot Club 108(3):388. https://doi.org/10.2307/2484725
Uzunova M, Ecke W (1999) Abundance, polymorphism and genetic mapping of microsatellites in oilseed rape (Brassica napus L.). Plant Breed 118:323–326
Varghese J, Rudolph B, Uzunova M, Ecke W (2000) Use of 5′-anchored primers for the enhanced recovery of specific microsatellite markers in Brassica napus L. Theor Appl Genet 101:115–119
Yeh F, Yang R, Boyle T, Ye Z, Mao J (2002) Popgen 32, microsoftware windows based freeware for population genetic analysis. Molecular Biology and Biotechnology Center, Edmonton
Yu C, Hu S, Zhao H, Guo A, Sun G (2005) Genetic distances revealed by morphological characters, isozymes, proteins and RAPD markers and their relationships with hybrid performance in oilseed rape (Brassica napus L.). Theor Appl Genet 110:511–518
Acknowledgements
We are very thankful to the members of Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture, Faisalabad, Pakistan for their support and encouragement to conduct this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
A. Raza, S.S. Mehmood, F. Ashraf, and R.S.A. Khan declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Raza, A., Mehmood, S.S., Ashraf, F. et al. Genetic Diversity Analysis of Brassica Species Using PCR-Based SSR Markers. Gesunde Pflanzen 71, 1–7 (2019). https://doi.org/10.1007/s10343-018-0435-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10343-018-0435-y