Abstract
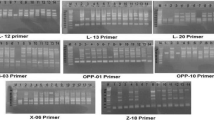
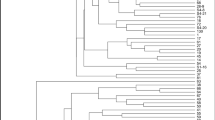
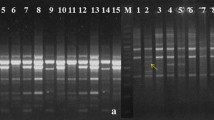
In recent years, plant breeding of some cultivars such as sumac has attracted more attention because their contributions to human health started to be revealed. Turkey being its origin, sumac (Rhus coriaria L.) is widely used as a medicinal plant and spice. However, the sporadic cultivation of cultivars such as sumac endangers its existence and future. Therefore, the protection of this cultivar and its genetic characterization as well as encouraging the cultivation of this cultivar through plant breeding is of utmost importance. In this respect, this study aims to determine the genetic diversities among 24 different sumac (Rhus coriaria L.) genotypes. 17 different SRAP (sequence related amplified polymorphism) and 12 ISSR (inter simple sequence repeat) primers were used in this study, and, as a result, 133 amplifications were obtained. 130 of these amplifications were polyphormic, the average value of polymorphism were determined as 97.74%. In addition, genetic difference among the genotypes were calculated, which yielded a percentage of 45%. The highest difference was observed between S19 and S5 (73%), while the lowest genetic difference was found between S23 and S24 genotypes (20%). These results suggest that sumac genotypes displaying high genetic difference will be selected as parents in the plant breeding studies on sumac cultivars in the future, thus increasing variation rate.


Similar content being viewed by others
References
Abdalla AM, Reddy OUK, El-Zik KM, Pepper AE (2001) Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor Appl Genet 102:222–229
Ateş Sönmezoğlu Ö, Yıldırım A, Eserkaya Gulec T, Kandemir N (2010) Using marker assisted selection in wheat breeding. Turk J Sci Res 3(2):67–79
Bardak A (2007) Genetic diversity of diploid and tetraploid cottons determined by SSR markers and its relationship with fiber quality traits. http://agris.fao.org/agris-search/search.do?recordID=TR2011001065
Bardak A, Bölek Y (2012) Genetic diversity of diploid and tetraploid cottons determined by SSR and ISSR markers. Turk J Field Crop 17(2):139–144
Başoğlu F, Cemeroğlu B (1984) Sumak’ın Kimyasal Bileşimi Üzerine Araştırma. Gıda Dergisi 9(3):167–172
Budak H, Shearman RC, Parmaksiz I, Dweikat I (2004) Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor Appl Genet 109(2):280–288
Çardaklı E, Bardak A, Özdemir M (2017) Determination of genetic diversity of some sage species collected from eastern mediterranean region. https://doi.org/10.24925/turjaf.v5i6.695-700.1183
Filiz E, Ozdemir BS, Budak F, Vogel JP, Tuna M, Budak H (2009) Molecular, morphological and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52(10):876–890
Gülşen O, Mutlu N (2005) Genetic markers used in plant sciences and their utilization. Alatarım 4(2):27–37
Gutierrez OS, Basu S, Saha S, Jenkins JN, Shoemaker DB, Cheatham CL, McCarty JC Jr (2002) Genetic distance among selected cotton genotypes and its relationship with F2 performance. Crop Sci 42:1841–1847
Kafkas S, Ozkan H, Ak BE, Acar I, Atli HS, Koyuncu S (2006) Detecting DNA polymorphism and genetic diversity in a wide pistachio germplasm: Comparison of AFLP, ISSR, and RAPD markers. J Am Soc Hortic Sci 131(4):522–529
Köroğlu A (1989) Difonognosic Studies on leaves and fruits of Rhus coriaria L. (Sumac) plant: Master dissertation. Ankara Institute of Health Sciences. 175 p
Kurucu S, Koyuncu M, Güvenç A, Başer KHC, Özek T (1993) The essential oils of Rhus coriaria L. (Sumac). J Essent Oil Res 5:481–486
Laborda PR, Oliveira KM, Garcia AA, Paterniani ME, De Souza AP (2005) Tropical maize germplasm: what can we say about its genetic diversity in the light of molecular markers. Theor Appl Genet 111(7):1288–1299
Li G, Quiros CF (2001) Sequence–related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. TAG. Theor Appl Genet 103(2):455–461
Liu S, Cantrell RG, McCarty JC Jr, Stewart JMD (2000) Simple sequence repeat based assessment of genetic diversity in cotton race stock accessions. Crop Sci 40(5):1459. https://doi.org/10.2135/cropsci2000.4051459x
Manjarrez-Sandoval P, Carter TE Jr, Webb DM, Burton JW (1997) RFLP genetic similarity and coefficient of parentage as genetic variance predictors for soybean yield. Crop Sci 37(3):698. https://doi.org/10.2135/cropsci1997.0011183X003700030002x
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Prakash S, Staden JV (2006) Assessment of genetic relationships between Rhus L. species using RAPD markers. Genet Resour Crop Evol 54:7–11. https://doi.org/10.1007/s10722-006-9007-6
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetic 155:945–959
Rahman M, Hussain D, Zafar Y (2002) Estimation of genetic divergence among elite cotton cultivars-genotypes by DNA fingerprinting technology. Crop Sci 42(6):2137. https://doi.org/10.2135/cropsci2002.2137
Rana MK, Bhat V (2005) RAPD markers for genetic diversity study among Indian cotton cultivars. Curr Sci 88:12–25
Rawashdeh IM, Nasri IH, Amri A (2009) Morphological diversity among and within populations and a medicinal plant (Achillea fragrantissima) collected from different regions of Jordan. Res Crop 37:242–252
Rayne S, Mazza G (2007) Biological activities of extracts from Sumac (Rhus L.) a review. Plant Foods Hum Nutr 62(4):165–175
Saebnazar A, Rahmani F (2013) Genetic variation among Salvia species based on sequence-related amplified polymorphism (SRAP) marker. J Plant Physiol Breed 38:108–1098
Senior LM, Murphy JP, Goodman MM, Stuber CV (1998) Utility of SSRs for determining genetic similarities and relationships in Maize using an agarose gel system. Crop Sci 38:1088–1098
Smith JSC, Smith OS (1992) Fingerprinting crop varieties. Adv Agron 47:85–140
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle JL (1997) An evaluation of the utility of SSR loci as molecular markers in Maize (Zea mays L.) comparisons with data from RLFP’s and pedigree. Theor Appl Genet 95:163–173
Sutyemez M (2016) New walnut Cultivars: Maras 18, Sutyemez 1, and Kaman 1. HortScience 51(10):1301–1303
Tabar VM, Chandrashekaran S, Rana MK, Bhaf KV (2004) RAPD analysis of genetic diversity in Indian tetraploid and diploid cotton (Gossypium spp). J Plant Biochem Biotechnol 13(1):81–84
Taşpınar MS, Tosun M (2002) Use of Isoenzyme electrophoretic technique in plant breeding. J Fac Agric 33(4):451–456
Wendel JF, Brubaker CL, Percival AE (1992) Genetic diversity in Gossypium hirsutum and the origin of Upland cotton. Am J Bot 79(11):1291–1310. https://doi.org/10.1002/j.1537-2197.1992.tb13734.x
Young ND (1996) QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol 34(1):479–501. https://doi.org/10.1146/annurev.phyto.34.1.479
Zhang JF, Lu Y, Adragna H, Hughs E (2005) Genetic improvement of New Mexico acala cotton gerplasm and their genetic diversity. Crop Sci 45(6):2363. https://doi.org/10.2135/cropsci2005.0140
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeats (SSR)-anchored PCR amplification. Genomics 20:176–183
Acknowledgements
This work was supported by Kahramanmaras Sutcu Imam University, Scientific Research Council (Project no:2017/1-25 YLS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
M. Sütyemez, G. Güvenç, Ş.B. Bükücü and A. Özcan declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Sütyemez, M., Güvenç, G., Bükücü, Ş.B. et al. The Determination of Genetic Diversity among some Sumac (Rhus coriaria L.) Genotypes. Erwerbs-Obstbau 61, 355–361 (2019). https://doi.org/10.1007/s10341-019-00440-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10341-019-00440-6