Abstract
Airway epithelial cells play an important role in the pathogenesis of inflammatory lung diseases such as asthma, cystic fibrosis and COPD. Studies concerning the function of the lipid metabolism of the airway epithelium are so far based only on the detection of lipids by immunohistochemistry but quantitative analyses have not been performed. Although recent advances in mass spectrometry have allowed to identify a variety of lipid classes simultaneously in isolated tissue samples, up until now, these methods were not suitable to analyze lipids in the airway epithelium. To determine all major lipid classes in airway epithelial cells, we used an LC–MS-based approach that can easily be combined with the specific isolation procedure to obtain epithelial cells. We tested the suitability of this method with a mouse model of experimental asthma. In response to allergen challenge, perturbations in the sphingolipids were detected, which led to increased levels of ceramides. We expanded the scope of this approach analysing human bronchus samples without pathological findings of adenocarcinoma patients. For the human lung epithelium an unusual lipid class distribution was found in which ceramide was the predominant sphingolipid. In summary, we show that disease progression and lipid metabolism perturbation can be monitored in animal models and that the method can be used for the analysis of clinical samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The role of lipids in pulmonary inflammatory diseases has gained increasing interest in recent years [1–6]. Previously, solely seen as essential components of membranes and energy sources, lipids are now regarded as important components of the immune response either by organizing signaling complexes in membranes such as lipid rafts [7] or by influencing the immune reaction by release of lipid-derived mediators such as arachidonic acid derived mediators [8] or sphingosine-1-phosphate [9].
Recent results indicate that changes in the lipid content of the airway epithelium play an important role in chronic airway diseases such as cystic fibrosis, COPD, and asthma. In cystic fibrosis, an increase in ceramide amounts was detected in the airway epithelium and was linked to inflammation, cell death and infection susceptibility [4]. Recent studies indicate that epithelial lipid metabolism is also changed in asthma [6, 10, 11]. For example, increased amounts of ceramides were detected in the airway epithelium of a guinea pig model as response to the induction of experimental allergic asthma [12]. Further support for an important role of lipid metabolism was found by genome wide association studies that identified the Orosomuid 1 like 3 (ORMDL3) gene to be strongly associated with childhood asthma [13]. ORMDL genes, which regulate ORM protein expression, are expressed in airway epithelial cells [14] and their expression is strongly increased in response to allergen challenge [15]. Functional studies have shown that the ORM families of proteins are regulators of the sphingolipid biosynthesis [16] pointing towards an important role of sphingolipids in the sensitization phase of asthma. Studies that identified the airway epithelium as cellular source of sphingolipids relied only on immunohistochemistry. Although immunohistochemistry is able to detect the cellular source of single lipids, only very limited information can be gathered regarding the amount of lipids as well as the lipid composition in these cells [12]. However, this information is crucial to understand the molecular mechanisms that underlie changes in lipid composition and possible cellular consequences of therapeutic interventions.
Recent advances in mass spectrometry have allowed identifying and quantifying a variety of lipid classes simultaneously. In the shotgun lipidomics approach lipid extracts are analyzed in direct infusion experiments solely relying on the performance of the tandem mass spectrometer utilized (MS/MS) [17–19]. This approach is fast and reliable for profiling lipidomes of a variety sample types [20–22]. However, lipid extracts containing considerable amounts of impurities, cannot be analyzed by shotgun lipidomics due to ion suppression effects. In such cases, liquid chromatography–mass spectrometry (LC–MS)-based methods are chosen [23–26]. The LC-separation as second analytical dimension is a must for the quantitation of very low abundant molecular species and lipid-derived signal molecules [27–29].
Here, we present a new lipidomics approach to determine the lipid content of airway epithelial cells, which was extensively tested in a mouse model of experimental asthma and applied to clinical samples. This method has previously been used to determine epithelial mediator mRNA expression [30] and was now optimized for lipid analysis using LC–MS. Epithelial cells were isolated with a sterile swab and was processed for the analysis by either normal phase (NP) or reversed phase (RP) LC–MS using high resolution Fourier transform ion cyclotron resonance mass spectrometry.
Experimental
Chemicals
All lipid standards with purities greater than 99 % were obtained from Avanti Polar Lipids (Alabaster, AL, USA). Deionized water was prepared with a Milli-Q purification system (Schwalbach, Germany); the conductivity of the water was less than 0.06 µS cm−1. All other chemicals (at least HPLC grade) were obtained from Sigma Aldrich (Traufkirchen, Germany) and used without further purification.
Animal Model of Allergic Airway Inflammation
Female Balb/c mice aged 8 weeks were treated intranasally with a crude extract of house dust mite (HDM) (Greer, Lenoir, NC, USA) in PBS (100 µg/50 µL) (n = 6) or PBS alone (n = 6) on days 0, 7, and 14. On day 15, the tracheae were removed and the epithelium was isolated as described below. Experiments were performed in two independent batches. All animals were held according to institutional guidelines with a 12 h day/night cycle and food and drink ad libitum. All animal experiments were approved by the Regierungspräsidium Giessen (Landgraf-Philipp-Platz 1-7, 35390 Gießen, Germany) and measures were taken to keep animal suffering to a minimum.
Isolation of Murine Tracheal Epithelium
To isolate the tracheal epithelium, the mice were killed by an overdose of isoflurane. The trachea was removed, transferred to a small cork plate with the tracheal muscle facing upwards and were fixed in position with insect needles. Hepes-Ringer solution was constantly applied to prevent drying of the tissue. Then, the tracheal muscle was cut using Vannas-Tübingen spring scissors and the tissue was flattened using additional insect needles. The epithelial surface was cleaned by rinsing repeatedly with HEPES-Ringer solution. Excess fluid was drained using a pipette without touching the epithelium. The epithelium was gently removed using a sterile swab (Single Polyester Fiber-Tipped Applicator Swab, Becton–Dickinson, Heidelberg Germany). The tip of the swab was cut with a sterile scalpel, transferred to a 1.5 mL Safe-Lock Eppendorf tube (Eppendorf AG, Hamburg, Germany), shock-frozen in liquid nitrogen and stored at −20 °C until the analysis.
Electron Microscopy
After removal of the murine epithelium, the trachea on the cork plate was transferred to a vial containing 2 % (v/v) glutardialdehyde, 0.6 % (w/v) paraformaldehyde, and 0.03 % (w/v) CaCl2 in 0.06 M cacodylate buffer, pH 7.3521 and fixed for at least 24 h. Then, the tissue was rinsed with cacodylate buffer, dehydrated in increasing concentrations of acetone, critical point dried, sputtered with gold and evaluated using a scanning electron microscope. Using ImageJ the total amount of epithelium removed from each trachea was calculated in 10 tracheae.
To further evaluate the procedure, tracheae from mice which were not treated intranasally were treated as described above but were processed for transmission electron microscopy after fixation. First, they were additionally fixed with osmium tetroxide, dehydrated with increasing concentrations of alcohol, transferred to propylene oxide and embedded in Araldite resin. Ultrathin sections were cut, transferred to grids, stained with lead citrate and uranyl acetate and evaluated by transmission electron microscopy.
Lipid Extraction of Murine Tracheal Epithelial Cells on Sterile Swabs
Mice tracheal epithelial cells on swabs were extracted according to Bligh and Dyer [31]. Briefly, 40 µL water were added to the swab and mixed with 100 µL of methanol and 50 µL of chloroform (sample/methanol/chloroform = 0.8/2/1 by volume). The mixture was vortexed and sonicated for 10 min. After 25 min at room temperature, 50 µL of water (1 volume) and 50 µL of chloroform (1 volume) were added, vortexed and centrifuged for 10 min at 400×g. The lower organic phase was transferred to another vial and the water phase was re-extracted two more times with 50 µL of chloroform (1 volume). Organic phases were pooled, solvent was removed in vacuum and samples were stored at −20 °C. Prior to analysis, dried samples were dissolved in 100 µL of chloroform/methanol = 86/13 (v/v) and 25 µL of the reconstituted sample was mixed with 25 µL of a mixture of deuterated lipid standards (1.43 ng µL−1 per species, consisting of 16:0 D31-18:1 PI, 16:0 D31 Cer, 16:0 D31-18:1 PG, 16:0 D31-18:1 PE, 16:0 PC D62, 16:0 D31-18:1 PS, 16:0 D31 SM).
Isolation of Human Bronchus Tissue Samples and Epithelial Cells
Human bronchial samples without pathological findings were obtained from lung lobes that were removed during surgery from one female and two male adenocarcinoma patients (age: 61–77 years). Pieces of the main bronchi were removed, cut open, fixed with needles on a cork plate and remaining mucus was removed. As described above for mouse tracheal epithelium, bronchial epithelial cells were removed with a sterile swab. In addition, whole bronchial tissue samples were also obtained. For each patient at least five epithelium samples were processed. After isolation, samples were extracted as described below. The use of patient materials was approved by the ethics committee of the University of Lübeck (AZ 12-220).
Lipid Extraction of Human Bronchus Tissue and Epithelium Samples
Prior to lipid extraction, human lung epithelial cells on swabs were moistened with 100 µL of water and human bronchus tissue samples were homogenized in a 20-fold excess of 50 mM KCl. From this tissue homogenate 50 µL were processed for lipid extraction, which corresponds to approximately, 2.5 mg of tissue sample. After the addition of 50 µL of the internal standard mixture (5 ng/µL per species, consisting of 16:0 D31-18:1 PI, 16:0 D31 Cer, 16:0 D31-18:1 PG, 16:0 D31-18:1 PE, 18:0-18:0 PC-OO, 16:0 D31-18:1 PS, 16:0 and D31 SM), lipids were extracted from the samples according to Bligh and Dyer [31]. Dried samples obtained after extraction were stored at −20 °C until LC–MS analysis. Prior to NP-LC–MS analysis, the residue was re-dissolved in 200 µL of chloroform/methanol = 86/13 (v/v). For RP-MS analysis, 50 µL of this solution were dried in vacuum and the dried residue was re-dissolved in 50 µL of methanol containing 0.1 % ammonium acetate.
Normal Phase Liquid Chromatography (NP-LC–MS)
An Agilent 1100 HPLC system (Agilent, Waldbronn, Germany) was used for the separation of the phospholipids on a 150 mm BETASIL Diol-100 column with a particle size of 5 µm (Thermo Fisher, Bremen, Germany) and 0.32 mm inner diameter using 5 μL injection volumes. The solvents, gradient profile and flow rates are summarized in supplementary Table 1.
Reversed Phase Liquid Chromatography (RP-LC–MS)
For the analysis of neutral lipids in the clinical samples, the Agilent 1100 HPLC system was used for the separation of the lipids by reversed phase HPLC on a 150 mm Biobasic C18 column (Thermo Fisher) with a 0.32 mm inner diameter using 5 μL injection volumes. The solvents, gradient profile and flow rates are summarized in supplementary Table 2.
Mass Spectrometry
All mass spectrometric analyses were performed on a high resolution Bruker Apex Qe FT-MS (Bruker Daltonics, Bremen, Germany) equipped with a 7 Tesla actively shielded magnet and an Apollo Dual ESI/MALDI ion source. For TAG analysis, full scan MS data were acquired in the first 40 min of the LC acquisition in the positive ion mode. For the phospholipid analysis, full scan MS data were acquired in the first 60 min of the LC-run. Instrumental parameters were as follows: source temperature at 200 °C, −3.8 and 4.5 kV ionization voltages for negative and positive ion mode analysis, respectively. For human lung samples 1 M data points sampling rate and 1 s accumulation time for each scan was used and the nebulizer gas was set to 1.0 L min−1, drying gas was set to 5 L min−1, and TOF time parameter was set to 0.0014. For mouse epithelium samples 512 k data points sampling rate and 0.6 s accumulation time for each scan was used, where 2 microscans were averaged; nebulizer gas was set to 1.3 L min−1, drying gas was set to 3 L min−1, and TOF time parameter was set to 0.0021. Data acquisition was performed using the HyStar 3.2 software (Bruker Daltonics).
Data Processing
NP-LC–MS and RP-LC- MS data files were processed using Bruker’s Data Analysis 4.0 software. Mass spectra were averaged in the retention time ranges of lipid classes of interest (defined by the elution time of the internal standards). Each mass spectrum was smoothed, baseline subtracted and finally peaks lists were extracted (m/z values and corresponding intensities). Lipids were identified by their monoisotopic masses with an accuracy better 5 ppm using LipidXplorer [32] and their respective retention time window. If available, intensity ratios of lipid species and their corresponding deuterated internal standard were used for quantification of lipids in the sample. DAG and TAG were quantified using 36:0 PC-OO as standard, which eluted closest to the elution time range of both lipid classes in the RP-LC–MS.
Data Analysis and Statistical Analysis
Statistical tests were performed using GraphPad Prism version 6.00 for Windows, (GraphPad Software, San Diego, CA, USA). A significant difference between lipid species in the two data sets was assumed if p < 0.05 for the Mann–Whitney U test.
Results
Selective Removal of Epithelial Cells from Murine Trachea Using a Sterile Swab
After trachea removal and rinsing the surface, the epithelial cells were obtained by gently skimming the epithelium using a sterile swab. To verify selective removal of epithelial cells from trachea samples, scanning electron microscopy and transmission electron microscopy experiments were performed. These experiments verified that the swab removed the epithelium without destroying the subepithelial dense fiber network and without reaching deeper parts of the tracheal tissue (Fig. 1). No remaining mucus or other material was detected on the remaining epithelial cells. As judged by the area of missing epithelium in the trachea we estimated that in average 5.23 ± 0.12 mm2 (n = 10) was removed using the swab. Such area would correspondent to approximately 80,000 cells, which subsequently were processed for lipid extraction and LC–MS analysis.
Electron microscopy of murine tracheae after epithelial removal. a, b Scanning electron micrograph of a murine trachea after removal of the epithelium with a sterile swab. a View of the whole trachea, arrows indicate holes from insect needles. b Magnification of the boxed area in a. The arrowheads indicate the border between removed epithelium (r) and remaining tracheal epithelium (e). c, d Transmission electron microscopy of an area with preserved (c) and removed (d) epithelium. Red arrows indicate the epithelial basement membrane
Analysis of Epithelial Lipids Using NP-LC–MS
Initially, we intended to analyse swab lipid extracts in a shotgun lipidomics approach allowing high-throughput sample analysis. Unfortunately, analyses without pre-separation were not successful due to contaminants from the swab plastic material, which gave rise to two major peaks at m/z 594.1605 and m/z 1170.2883 in the positive ion mode. These contaminants made a sensitive lipid quantitation impossible due to strong ionization suppression. Note, several other swabs made from different material were tested but showed even higher chemical background. To overcome these issues and to allow automated data analysis using LipidXplorer, an NP-LC–MS method was implemented.
The sensitivity and reproducibility of the NP-LC–MS method for phospholipid analysis was estimated using a lipid standard mixture containing Cer, PG, PE, PI, PS, SM, and PC. The limit of detection (LOD) for all lipids was in the range of some hundred fmol on column (supplementary Table 3). For PS the LOD was in the low pmol range as this lipid class elutes very late under the chosen chromatographic conditions resulting in peak broadening and reduced signal intensity in the resulting mass spectra.
To identify and quantify lipids using NP-LC–MS analysis, all mass spectra in the elution time range of a specific lipid class of interest were averaged. Thus, retention time stability is of crucial importance for automated data analysis. Retention time stability was determined from 25 subsequent analyses for the three standard lipids eluting at different retention times (16:0 D31 Cer at 1.2 min, D31-18:1 PE at 14 min, 16:0 D31-18:1 PS at 32 min) and found to have only relative standard deviation of 5 %. It should be noted that retention time ranges used for quantification of phospholipids were adjusted for each batch of samples analysed as retention time discrepancies were observed between different column batches and mobile phase preparations. In our study, sets of up to 30 epithelium samples were measured and mass spectra for specific lipid classes could automatically be processed because the retention times within one batch remained stable despite matrix effects (see supplementary Fig. 1). The established workflow for the analysis of lipids from airway epithelial cells is summarized in Fig. 2a. Epithelial cells are removed from the trachea using a sterile swab and lipids are extracted using standard extraction procedures. Extracts are analysed using NP-LC to separate chemical contaminants of the swabs from the lipids. High resolution ESI-FT-ICR-MS spectra were acquired online and within defined retention time windows for each lipid class one mass spectrum was obtained (Fig. 2b). Peak lists were computed with standard spectra processing tools, which were used as input for lipid identification using the LipidXplorer software. Lipid quantitation was achieved using the response of an internal standard that co-eluted with the lipid class of interest.
a Workflow for the analysis of lipids in murine and human lung epithelial cells. Epithelial cells are removed from trachea samples using a sterile swab. The tip of the swab is subsequently extracted using chloroform/methanol and measured using NP-µHPLC-ESI FTICRMS in the positive and negative ion mode. Mass spectra are averaged in the retention time ranges of lipid classes of interest (defined by the elution time of the internal standards), smoothed, baseline subtracted and peaks lists (m/z values and corresponding intensities) are exported for each mass spectrum. Lipids in the mass spectra were identified by their monoisotopic masses (5 ppm accuracy) and their retention time using LipidXplorer. b Typical LC-MS base peak ion chromatogram obtained from epithelium cells (upper left) and exemplified mass spectra obtained after averaging mass ranges used for identification of PI (upper right), PS (lower left), and PG (lower right). (Asterisk unknown compound not observed in blank controls)
Sensitive Analysis of Ceramide Molecular Species in the Negative Ion Mode
For the analysis of lipids extracted from murine tracheal epithelial cells, a chloroform-based solvent system was used for separation of lipids on a normal phase column [25]. Use of this solvent system favors formation of [M + Cl]− ions of ceramides while the [M-H]− ions are barely detectable in the negative ion mode as is shown in supplementary Fig. 2a. In the positive ion mode, [M + H]+ ions of ceramides are formed (supplementary Fig. 2b) but with comparably low ionization efficiency. (supplementary Fig. 2a, b). In addition, swab contaminants co-eluted with ceramides and were detected at high abundance in the positive ion mode. As a result of these observations, we choose to quantify ceramide species as chloride adducts in the negative ion mode, which was also reported earlier [33–35].
Lipid Composition of Murine Tracheal Epithelial Cells and Changes due to Induction of Experimental Asthma
In the present work, we analysed lipids in epithelial cells obtained directly from murine trachea. From approximately, 80000 cells collected, 99 lipid species belonging to seven different lipid classes were identified and quantified in pmol/sample. SM and PC were found to be the major components of epithelial cells of the mouse trachea with approximately 68 molpercent (PC and PC-O: 41 %, SM: 26 %) followed by PE and PE-O (19 %). As minor components of the membrane lipids, PI (6 %), PS (4 %), PG (1 %) and Cer (1 %) were identified (supplementary Table 4).
Sphingolipid metabolic perturbations were recently implicated as important risk factors for childhood asthma [36]. In this regard, we tested if our method could help to assess lipid metabolic changes in the epithelia between house dust mite challenged mice (HDM) and control animals (PBS) (Fig. 3). Here we could observe that ceramides were predominately changed on the lipid class level after HDM challenge. A closer look into the ceramide profile revealed that all species after HDM challenge were increased (Fig. 3 inset). Further prominent changes were found for PC-O 32:0, PC-O 34:1 and PS 34:1 that are significantly increased in HDM challenged animals, whereas PG 34:2 and PG 32:1 are significantly decreased when compared to control mice (supplementary Table 4, p < 0.05, Mann–Whitney test).
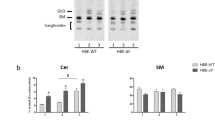
Lipid class distribution in pmoles/sample determined in murine tracheal epithelial cells of animals challenged using house dust mite extract (red) and healthy controls (blue). The inset shows the single species distribution of ceramide species normalized to control samples (PBS). Bar graphs (main panel) and scatter dot plots (inset) show mean ± SD. Significantly changed species (Mann–Whitney U test) are marked with asterisks: *p < 0.05; **p < 0.01; ***p < 0.001
The Lipidome of Human Bronchus Epithelial Cells
Encouraged by the results of the mouse model we tested whether our method could in principle also be applied for the human bronchial epithelium. Subsequently, epithelium cell samples were obtained from different locations of explanted bronchial tissues of confirmed adenocarcinoma patients that did not show histological alterations in the excised bronchi. The abundance of selected lipid classes in the bronchus epithelial cells obtained from three different patients is shown in Fig. 4. For the analysis of the human airway epithelial cells approximately, a tenfold higher amount of biological material can be obtained as compared to the murine trachea. That further resulted in an increase of lipid identifications to approximately, 300 lipids (supplementary Table 5). For all patients, epithelium samples were isolated on at least five locations of the bronchus. It further enabled us to determine the variability of lipid profiles for the three individuals. For all patients, PC was found to be the most abundant lipid class followed by PE plus PE-O. For patient 2 a high variability compared to the other patients was detected in the lipid content of TAG, SM and Cer, which cannot be attributed to a specific functional or clinical parameter. In future studies, the number of patients has to be increased to access the inter- and intra-individual lipidome variability to draw functional associations. We further tested the specificity of the isolation technique by comparing whole bronchial tissue samples with isolated epithelium samples (Table 1). On level of the lipid class distribution the strongest difference can be found in reduced TAG amounts in the epithelium, which is related to peribronchial adipose tissue. We further observed a clear increase in abundance for Cer, PE-O and PE levels in the epithelium (Table 1). A closer look into the Cer and SM profiles revealed a shift towards shorter acyl chains for the epithelium samples (Fig. 5 a, b). This effect is more prominent for the ceramides where Cer 34:1 abundance is increased by a factor of 2 compared to the whole bronchus. It is further an important observation that Cer is the predominant sphingolipid in the epithelium, which was not expected.
Lipid class distribution for selected lipid classes in of human airway epithelial cells obtained from three different patients. N = 3 patients, at least five samples per patient were analyzed, each dot corresponds to one swab containing epithelial cells, which was extracted and analyzed using the workflow shown in Fig. 2
Sphingolipid profiles of human bronchus. a Ceramide (Cer) and b sphingomyelin (SM) single species distribution (mol % of total identified Cer and SM, respectively) of human bronchial epithelial cells and bronchus tissue samples. Epithelium samples: n = 3 patients, at least five samples per patient were analyzed; bronchus tissue samples: n = 3 patients, at least two samples per patient were analyzed. Bar graphs show mean ± SD
Lipid Profiles of Mouse and Human Airway Epithelium
For many respiratory diseases are mouse models established, which help to investigate fundamental processes in disease progression [37–40]. Here we present the first comparison of the lipid composition of mouse and human airway epithelium. PC was in both epithelia the most abundant membrane lipid (Fig. 6a). A striking difference was observed in the abundance of SM, which in the mice is as nearly abundant as PC. For the human epithelium we observed that Cer is the most abundant sphingolipid and approximately, tenfold more abundant compared to the mouse. Further changes were found for the PC-O content, which was significantly decreased in the human airway epithelium whereas for PG a slight increase of 25 % was observed. In context of the importance of sphingolipids we like to underline that the acyl chain distribution in ceramides as well as sphingomyelin is very different (Fig. 6b, c) between mouse and human airway epithelium. For the human as well mouse airway epithelium the Cer 34:1 and SM 34:1 are the predominant species. But long chain sphingolipids make up a higher proportion in the mouse epithelium, which can be best recognized for Cer 40:1, Cer 42:1 and SM 42:2. The functional impact of such global lipid changes is until now out of our reach, but might be of importance in the future when results of animal models are translated for clinical application.
Lipid profiles of murine and human airway epithelial cells. a Membrane lipid class distribution, b single species distribution of even-numbered ceramide species identified, c single species distribution of even-numbered sphingomyelin species identified. Bar graphs show mean ± SD; significantly changed species (Mann–Whitney U test) are labeled with asterisks: *p < 0.05; ****p < 0.0001
Discussion
A method was developed to investigate the lipid composition of lung epithelia using a sterile swab for cell isolation and LC–MS for quantitation. The approach was tested in a mouse model of experimental asthma and human samples. Even though standard plastic consumable were utilized for the isolation of the epithelia this method allowed quantifying many important membrane lipids. In this way, a molecular snapshot of the lipid metabolism of the airway epithelium can be derived, which will help to investigate the influence of the lipid metabolism on lung diseases. To our knowledge, we report for the first time the lipid profiles of phospholipids, sphingolipids and neutral lipids of airway epithelial cells.
Our method is sensitive enough to quantify ceramide species in the airway epithelium. In general, ceramides and sphingolipids are implicated as key molecules in the development of asthma and inflammatory processes [6, 11, 36]. Our observation of increased ceramide amounts in allergen-challenged mice supports this finding. This view is also supported by a recent study in which inhibition of Cer synthase resulted in a decrease of Cer levels in lung tissue samples in a guinea pig model of experimental asthma and reduced asthmatic hallmarks in challenged animals [12].
For the analysis of intrapulmonary airway epithelium of mice, however, our technique will be of limited use due to the large size of the sterile swab. To improve the depth of lipidomics studies, other techniques to isolate the epithelium have to be found. The use of laser capture microdissection can be a promising approach to study lung tissue sections [41] as well as MALDI-based mass spectrometric imaging [42]. To further enhance the impact of such studies we will expand our approach to LC–MSn-based techniques that enable us to measure secondary messenger like lipid mediators, sphingosines and lyso-lipids [43]. We have shown that this method is also applicable to specifically analyze the lipid composition of human epithelial samples obtained from explanted lung samples. Since epithelial brushings can be performed by bronchoscopy, our method could be a useful tool to study the lipid metabolism of the human airway epithelium of patients.
Conclusions
We developed a LC–MS-based method that can become a valuable tool to investigate the role of lipids in the airway epithelium in context of respiratory diseases. The approach is applicable for mouse model as well as to profile lipids of the human airway epithelium. Our method can in principle be used to analyze lipid profiles of epithelium cells obtained by epithelial brushings performed during bronchoscopy. We see here the possibility to utilize this platform technology for clinical application.
Abbreviations
- NP-LC–MS:
-
Normal phase liquid chromatography mass spectrometry
- RP-LC–MS:
-
Reversed phase liquid chromatography mass spectrometry
- Cer:
-
Ceramides
- PG:
-
Phosphatidylglycerol
- SM:
-
Sphingomyelin
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- PE-O:
-
Ether phosphatidylethanolamine
- PI:
-
Phosphatidylinositol
- CL:
-
Cardiolipin
- PS:
-
Phosphatidylserine
- PC-O:
-
Ether phosphatidylcholine
- PC-OO:
-
Diether phosphatidylcholine
- TAG:
-
Triacylglycerol
- DAG:
-
Diacylglycerol
References
Garcia-Sastre A (2013) Lessons from lipids in the fight against influenza. Cell 154(1):22–23. doi:10.1016/j.cell.2013.06.024
Levy BD (2013) Sphingolipids and susceptibility to asthma. New Engl J Med 369(10):976–978. doi:10.1056/NEJMcibr1306864
Scarpa MC, Baraldo S, Marian E, Turato G, Calabrese F, Saetta M, Maestrelli P (2013) Ceramide expression and cell homeostasis in chronic obstructive pulmonary disease. Res Int Rev Thorac Dis 85(4):342–349. doi:10.1159/000341185
Teichgräber V, Ulrich M, Endlich N, Riethmuller J, Wilker B, De Oliveira-Munding CC, van Heeckeren AM, Barr ML, von Kurthy G, Schmid KW, Weller M, Tummler B, Lang F, Grassme H, Doring G, Gulbins E (2008) Ceramide accumulation mediates inflammation, cell death and infection susceptibility in cystic fibrosis. Nat Med 14(4):382–391. doi:10.1038/nm1748
Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, Silver RB, Jiang XC, Worgall S (2013) Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med 5(186):186ra167. doi:10.1126/scitranslmed.3005765
Yang Y, Uhlig S (2011) The role of sphingolipids in respiratory disease. Ther Adv Res Dis 5(5):325–344. doi:10.1177/1753465811406772
Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1(1):31–39. doi:10.1038/35036052
Farooqui AA (2009) Lipid mediators in the neural cell nucleus: their metabolism, signaling, and association with neurological disorders. Neurosci Rev J Bring Neurobiol Neurol Psychiatry 15(4):392–407. doi:10.1177/1073858409337035
Spiegel S, Milstien S (2011) The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol 11(6):403–415. doi:10.1038/nri2974
Nixon GF (2009) Sphingolipids in inflammation: pathological implications and potential therapeutic targets. Br J Pharmacol 158(4):982–993. doi:10.1111/j.1476-5381.2009.00281.x
Uhlig S, Gulbins E (2008) Sphingolipids in the lungs. Am J Respir Crit Care Med 178(11):1100–1114. doi:10.1164/rccm.200804-595SO
Masini E, Giannini L, Nistri S, Cinci L, Mastroianni R, Xu W, Comhair SA, Li D, Cuzzocrea S, Matuschak GM, Salvemini D (2008) Ceramide: a key signaling molecule in a Guinea pig model of allergic asthmatic response and airway inflammation. J Pharmacol Exp Ther 324(2):548–557. doi:10.1124/jpet.107.131565
Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448(7152):470–473. doi:10.1038/nature06014
Roeder T, Isermann K, Kabesch M (2009) Drosophila in asthma research. Am J Respir Crit Care Med 179(11):979–983. doi:10.1164/rccm.200811-1777PP
Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, Rosenthal P, Mueller JL, Hoffman HM, Suzukawa M, Niwa M, Broide DH (2012) ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA 109(41):16648–16653. doi:10.1073/pnas.1204151109
Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463(7284):1048–1053. doi:10.1038/nature08787
Schwudke D, Schuhmann K, Herzog R, Bornstein SR, Shevchenko A (2011) Shotgun lipidomics on high resolution mass spectrometers. Cold Spring Harb Perspect Biol 3(9):a004614. doi:10.1101/cshperspect.a004614
Yang K, Cheng H, Gross RW, Han X (2009) Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem 81(11):4356–4368. doi:10.1021/ac900241u
Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A (2006) Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem 78(17):6202–6214. doi:10.1021/ac060545x
Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A (2011) Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci USA 108(5):1903–1907. doi:10.1073/pnas.1019267108
Carvalho M, Sampaio JL, Palm W, Brankatschk M, Eaton S, Shevchenko A (2012) Effects of diet and development on the Drosophila lipidome. Mol Syst Biol 8:600. doi:10.1038/msb.2012.29
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49(5):1137–1146. doi:10.1194/jlr.D700041-JLR200
Hutchins PM, Barkley RM, Murphy RC (2008) Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J Lipid Res 49(4):804–813. doi:10.1194/jlr.M700521-JLR200
McLaren DG, Miller PL, Lassman ME, Castro-Perez JM, Hubbard BK, Roddy TP (2011) An ultraperformance liquid chromatography method for the normal-phase separation of lipids. Anal Biochem 414(2):266–272. doi:10.1016/j.ab.2011.03.009
Hubner G, Crone C, Lindner B (2009) lipID–a software tool for automated assignment of lipids in mass spectra. J Mass Spectrom JMS 44(12):1676–1683. doi:10.1002/jms.1673
Nakanishi H, Ogiso H, Taguchi R (2009) Qualitative and quantitative analyses of phospholipids by LC-MS for lipidomics. Methods Mol Biol 579:287–313. doi:10.1007/978-1-60761-322-0_15
Whitehead SN, Hou W, Ethier M, Smith JC, Bourgeois A, Denis R, Bennett SA, Figeys D (2007) Identification and quantitation of changes in the platelet activating factor family of glycerophospholipids over the course of neuronal differentiation by high-performance liquid chromatography electrospray ionization tandem mass spectrometry. Anal Chem 79(22):8539–8548. doi:10.1021/ac0712291
O’Donnell VB (2011) Mass spectrometry analysis of oxidized phosphatidylcholine and phosphatidylethanolamine. Biochim Biophys Acta 1811(11):818–826. doi:10.1016/j.bbalip.2011.07.018
Saigusa D, Shiba K, Inoue A, Hama K, Okutani M, Iida N, Saito M, Suzuki K, Kaneko T, Suzuki N, Yamaguchi H, Mano N, Goto J, Hishinuma T, Aoki J, Tomioka Y (2012) Simultaneous quantitation of sphingoid bases and their phosphates in biological samples by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Bioanal Chem 403(7):1897–1905. doi:10.1007/s00216-012-6004-9
Bermbach S, Weinhold K, Roeder T, Petersen F, Kugler C, Goldmann T, Rupp J, Konig P (2014) Mechanisms of cilia-driven transport in the airways in the absence of mucus. Am J Respir Cell Mol Biol 51(1):56–67. doi:10.1165/rcmb.2012-0530OC
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8):911–917
Herzog R, Schuhmann K, Schwudke D, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2012) LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One 7(1):e29851. doi:10.1371/journal.pone.0029851
Kuksis A, Marai L, Myher JJ (1991) Plasma lipid profiling by liquid chromatography with chloride-attachment mass spectrometry. Lipids 26(3):240–246
Hsu FF, Turk J (2002) Characterization of ceramides by low energy collisional-activated dissociation tandem mass spectrometry with negative-ion electrospray ionization. J Am Soc Mass Spectrom 13(5):558–570. doi:10.1016/S1044-0305(02)00358-6
Zhu J, Cole RB (2000) Formation and decompositions of chloride adduct ions. J Am Soc Mass Spectrom 11(11):932–941
Maceyka M, Spiegel S (2014) Sphingolipid metabolites in inflammatory disease. Nature 510(7503):58–67. doi:10.1038/nature13475
Starkey MR, Jarnicki AG, Essilfie AT, Gellatly SL, Kim RY, Brown AC, Foster PS, Horvat JC, Hansbro PM (2013) Murine models of infectious exacerbations of airway inflammation. Curr Opin Pharmacol 13(3):337–344. doi:10.1016/j.coph.2013.03.005
Chapman DG, Tully JE, Nolin JD, Jansen-Heininger YM, Irvin CG (2014) Animal models of allergic airways disease: where are we and where to next? J Cell Biochem. doi:10.1002/jcb.24881
Fricker M, Deane A, Hansbro PM (2014) Animal models of chronic obstructive pulmonary disease. Expert Opin Drug Discov 9(6):629–645. doi:10.1517/17460441.2014.909805
Zhou Z, Duerr J, Johannesson B, Schubert SC, Treis D, Harm M, Graeber SY, Dalpke A, Schultz C, Mall MA (2011) The ENaC-overexpressing mouse as a model of cystic fibrosis lung disease. J Cyst Fibros Off J Eur Cyst Fibros Soc 10(Suppl 2):S172–S182. doi:10.1016/S1569-1993(11)60021-0
Hebbar S, Schulz WD, Sauer U, Schwudke D (2014) Laser capture microdissection coupled with on-column Extraction LC-MSn enables lipidomics of fluorescently labeled drosophila neurons. Anal Chem 86(11):5345–5352. doi:10.1021/ac500276r
Murphy RC, Hankin JA, Barkley RM (2009) Imaging of lipid species by MALDI mass spectrometry. J Lipid Res 50(Suppl):S317–S322. doi:10.1194/jlr.R800051-JLR200
Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, Sullards MC, Merrill AH Jr (2009) Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res 50(8):1692–1707. doi:10.1194/jlr.D800051-JLR200
Acknowledgments
The financial support by the DFG (SFB TR22 Z01 to BL; SFB TR22 Z06 to DS; TR22 Z05 to PK) and the German Ministry for Education and Research (82DZL00102 to PK) is gratefully acknowledged. Human lung tissue samples were obtained by the BioMaterialBank Nord, which is funded in part by the Airway Research Center North (ARCN), Member of the German Center for Lung Research and is member of the popgen 2.0 network (P2N), which is supported by a grant from the German Ministry for Education and Research (01EY1103). We thank Dr. med Christian Kugler (Großhansdorf Hospital) for providing human tissue samples. We thank Gudrun Knebel for expert technical assistance.
Conflict of interest
All authors have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Published in the topical collection Recent Developments in Clinical Omics with guest editors Martin Giera and Manfred Wuhrer.
Lipid species are abbreviated as follows: [abbreviation of the lipid class] [sum of carbon atoms in all fatty acid residues]:[number of double bonds], e.g. PC 36:1 describes phosphatidylcholine containing 36 carbon atoms and one double bond in the fatty acid chains.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10337_2014_2787_MOESM5_ESM.pdf
Supplementary Tab. 5 Lipid species distribution in human bronchus tissue samples and airway epithelial cells (PDF 808 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zehethofer, N., Bermbach, S., Hagner, S. et al. Lipid Analysis of Airway Epithelial Cells for Studying Respiratory Diseases. Chromatographia 78, 403–413 (2015). https://doi.org/10.1007/s10337-014-2787-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-014-2787-5