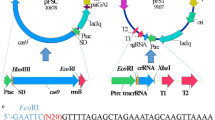
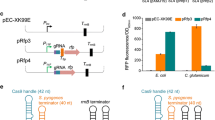
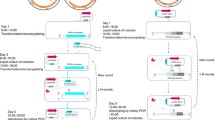
Abstract
Corynebacterium glutamicum is an essential industrial strain that has been widely harnessed for the production of all kinds of value-added products. Efficient multiplex gene editing and large DNA fragment deletion are essential strategies for industrial biotechnological research. Cpf1 is a robust and simple genome editing tool for simultaneous editing of multiplex genes. However, no studies on effective multiplex gene editing and large DNA fragment deletion by the CRISPR/Cpf1 system in C. glutamicum have been reported. Here, we developed a multiplex gene editing method by optimizing the CRISPR/Cpf1-RecT system and a large chromosomal fragment deletion strategy using the CRISPR/Cpf1-RecET system in C. glutamicum ATCC 14067. The CRISPR/Cpf1-RecT system exhibited a precise editing efficiency of more than 91.6% with the PAM sequences TTTC, TTTG, GTTG or CTTC. The sites that could be edited were limited due to the PAM region and the 1–7 nt at the 5′ end of the protospacer region. Mutations in the PAM region increased the editing efficiency of the − 6 nt region from 0 to 96.7%. Using a crRNA array, two and three genes could be simultaneously edited in one step via the CRISPR/Cpf1-RecT system, and the efficiency of simultaneously editing two genes was 91.6%, but the efficiency of simultaneously editing three genes was below 10%. The editing efficiency for a deletion of 1 kb was 79.6%, and the editing efficiencies for 5- and 20 kb length DNA fragment deletions reached 91.3% and 36.4%, respectively, via the CRISPR/Cpf1-RecET system. This research provides an efficient and simple tool for C. glutamicum genome editing that can further accelerate metabolic engineering efforts and genome evolution.




Similar content being viewed by others
References
Brooks AK, Gaj T (2018) Innovations in CRISPR technology. Curr Opin Biotechnol 52:95–101. https://doi.org/10.1016/j.copbio.2018.03.007
Cho JS, Choi KR, Prabowo CPS, Shin JH, Yang D, Jang J, Lee SY (2017) CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab Eng 42:157–167. https://doi.org/10.1016/j.ymben.2017.06.010
Choi JW, Jeon EJ, Jeong KJ (2019) Recent advances in engineering Corynebacterium glutamicum for utilization of hemicellulosic biomass. Curr Opin Biotechnol 57:17–24. https://doi.org/10.1016/j.copbio.2018.11.004
Cleto S, Jensen JV, Wendisch VF, Lu TK (2016) Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol 5:375–385. https://doi.org/10.1021/acssynbio.5b00216
Ellis HM, Yu D, DiTizio T (2001) High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA 98:6742–6746. https://doi.org/10.1073/pnas.121164898
Huang Y, Li L, Xie S, Zhao N, Han S, Lin Y, Zheng S (2017) Recombineering using RecET in Corynebacterium glutamicum ATCC14067 via a self-excisable cassette. Sci Rep 7:7916. https://doi.org/10.1038/s41598-017-08352-9
Jakočiūnas T, Bonde I, Herrgård M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28:213–222. https://doi.org/10.1016/j.ymben.2015.01.008
Jiang Y, Qian F, Yang J, Liu Y, Dong F, Xu C, Sun B, Chen B, Xu X, Li Y (2017) CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat Commun 8:15179. https://doi.org/10.1038/ncomms15179
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Lee J-Y, Na Y-A, Kim E, Lee H-S, Kim P (2016) The actinobacterium Corynebacterium glutamicum, an industrial workhorse. J Microbiol Biotechnol 26:807–822. https://doi.org/10.4014/jmb.1601.01053
Li L, Wei K, Zheng G, Liu X, Chen S, Jiang W, Lu Y (2018) CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl Environ Microbiol 84:e00827–e1818. https://doi.org/10.1128/AEM.00827-18
Li M, Chen J, Wang Y, Liu J, Huang J, Chen N, Zheng P, Sun J (2020) Efficient multiplex gene repression by CRISPR-dCpf1 in Corynebacterium glutamicum. Front Bioeng Biotechnol 8:357. https://doi.org/10.3389/fbioe.2020.00357
Liu J, Wang Y, Lu Y, Zheng P, Sun J, Ma YJMcf, (2017) Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb Cell Fact 16:205. https://doi.org/10.1186/s12934-017-0815-5
Oh J-H, van Pijkeren J-PJN (2014) CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42:e131–e131. https://doi.org/10.1093/nar/gku623
Okibe N, Suzuki N, Inui M, Yukawa H (2011) Efficient markerless gene replacement in Corynebacterium glutamicum using a new temperature-sensitive plasmid. J Microbiol Methods 85:155–163. https://doi.org/10.1016/j.mimet.2011.02.012
Su T, Liu F, Gu P, Jin H, Chang Y, Wang Q, Liang Q, Qi QJSr, (2016) A CRISPR-Cas9 assisted non-homologous end-joining strategy for one-step engineering of bacterial genome. Sci Rep 6:37895. https://doi.org/10.1038/srep37895
Sun B, Yang J, Yang S, Ye RD, Chen D, Jiang YJB (2018) A CRISPR-Cpf1-assisted non-homologous end joining genome editing system of Mycobacterium smegmatis. Biotechnol J 13:1700588. https://doi.org/10.1002/biot.201700588
Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa HJAEM (2005) New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480. https://doi.org/10.1128/AEM.71.12.8472-8480.2005
Świat MA, Dashko S, den Ridder M, Wijsman M, van der Oost J, Daran J-M, Daran-Lapujade P (2017) Fn Cpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res 45:12585–12598. https://doi.org/10.1093/nar/gkx1007
Wang B, Hu Q, Zhang Y, Shi R, Chai X, Liu Z, Shang X, Zhang Y, Wen T (2018) A RecET-assisted CRISPR–Cas9 genome editing in Corynebacterium glutamicum. Microb Cell Fact 17:63. https://doi.org/10.1186/s12934-018-0910-2
Wang M, Mao Y, Lu Y, Tao X, J-kJMp Z (2017) Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant 10:1011–1013. https://doi.org/10.1016/j.molp.2017.03.001
Wang Y, Liu Y, Liu J, Guo Y, Fan L, Ni X, Zheng X, Wang M, Zheng P, Sun J, Ma Y (2018) MACBETH: multiplex automated Corynebacterium glutamicum base editing method. Metab Eng 47:200–210. https://doi.org/10.1016/j.ymben.2018.02.016
Wu J, Deng A, Sun Q, Bai H, Sun Z, Shang X, Zhang Y, Liu Q, Liang Y, Liu S, Che Y (2017) Bacterial genome editing via a designed toxin–antitoxin cassette. ACS Synth Biol 7:822–831. https://doi.org/10.1021/acssynbio.6b00287
Yan M-Y, Yan H-Q, Ren G-X, Zhao J-P, Guo X-P, Sun Y-CJAEM (2017) CRISPR-Cas12a-assisted recombineering in bacteria. Appl Environ Microbiol 83:e00947–e1917. https://doi.org/10.1128/AEM.00947-17
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev AJC (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. https://doi.org/10.1016/j.cell.2015.09.038
Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JSJNb, (2017) Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol 35:31. https://doi.org/10.1038/nbt.3737
Zhang J, Yang F, Yang Y, Jiang Y, Huo Y-X (2019) Optimizing a CRISPR-Cpf1-based genome engineering system for Corynebacterium glutamicum. Microb Cell Fact 18:60. https://doi.org/10.1186/s12934-019-1109-x
Acknowledgements
All the authors are thankful for the financial support of the National Natural Science Foundation of China (Grant No. 31671840), the National Key R&D Program of China (Grant No. 2018YFA0901700) and the China Postdoctoral Science Foundation (Grant No. 2019M661676).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, N., Li, L., Luo, G. et al. Multiplex gene editing and large DNA fragment deletion by the CRISPR/Cpf1-RecE/T system in Corynebacterium glutamicum. J Ind Microbiol Biotechnol 47, 599–608 (2020). https://doi.org/10.1007/s10295-020-02304-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02304-5