Abstract
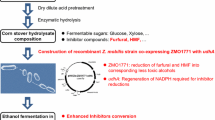
Furfural is a toxic by-product formulated from pretreatment processes of lignocellulosic biomass. In order to utilize the lignocellulosic biomass on isobutanol production, inhibitory effect of the furfural on isobutanol production was investigated and combinatorial application of two oxidoreductases, FucO and YqhD, was suggested as an alternative strategy. Furfural decreased cell growth and isobutanol production when only YqhD or FucO was employed as an isobutyraldehyde oxidoreductase. However, combinatorial overexpression of FucO and YqhD could overcome the inhibitory effect of furfural giving higher isobutanol production by 110 % compared with overexpression of YqhD. The combinatorial oxidoreductases increased furfural detoxification rate 2.1-fold and also accelerated glucose consumption 1.4-fold. When it compares to another known system increasing furfural tolerance, membrane-bound transhydrogenase (pntAB), the combinatorial aldehyde oxidoreductases were better on cell growth and production. Thus, to control oxidoreductases is important to produce isobutanol using furfural-containing biomass and the combinatorial overexpression of FucO and YqhD can be an alternative strategy.




Similar content being viewed by others
References
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. doi:10.1038/nature06450
Atsumi S, Li Z, Liao JC (2009) Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl Environ Microbiol 75:6306–6311. doi:10.1128/AEM.01160-09
Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC (2010) Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol 85:651–657. doi:10.1007/s00253-009-2085-6
Baek JM, Mazumdar S, Lee SW, Jung MY, Lim JH, Seo SW, Jung GY, Oh MK (2013) Butyrate production in engineered Escherichia coli with synthetic scaffolds. Biotechnol Bioeng 110:2790–2794. doi:10.1002/bit.24925
Barciszewski J, Siboska GE, Pedersen BO, Clark BF, Rattan SI (1997) A mechanism for the in vivo formation of N6-furfuryladenine, kinetin, as a secondary oxidative damage product of DNA. FEBS Lett 414:457–460
Boronat A, Aguilar J (1979) Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J Bacteriol 140:320–326
Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60:165–182. doi:10.1146/annurev.arplant.043008.092125
Gonzalez-Benito G, Rodriguez-Brana L, Bolado S, Coca M, Garcia-Cubero MT (2009) Batch ethanol fermentation of lignocellulosic hydrolysates by Pichia stipitis. Effect of acetic acid, furfural and HMF. New Biotechnol 25:S261
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349. doi:10.1007/s00253-005-0142-3
Gutierrez T, Ingram LO, Preston JF (2006) Purification and characterization of a furfural reductase (FFR) from Escherichia coli strain LYO1—an enzyme important in the detoxification of furfural during ethanol production. J Biotechnol 121:154–164. doi:10.1016/j.jbiotec.2005.07.003
Horvath IS, Taherzadeh MJ, Niklasson C, Liden G (2001) Effects of furfural on anaerobic continuous cultivation of Saccharomyces cerevisiae. Biotechnol Bioeng 75:540–549
Jarboe LR (2011) YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 89:249–257. doi:10.1007/s00253-010-2912-9
Jarboe LR, Liu P, Kautharapu KB, Ingram LO (2012) Optimization of enzyme parameters for fermentative production of biorenewable fuels and chemicals. Comput Struct Biotechnol J 3:e201210005. doi:10.5936/csbj.201210005
Khan QA, Shamsi FA, Hadi SM (1995) Mutagenicity of furfural in plasmid DNA. Cancer Lett 89:95–99
Kumar P, Barrett DM, Delwiche MJ, Stroeve P (2009) Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res 48:3713–3729. doi:10.1021/Ie801542g
Liu X, Bastian S, Snow CD, Brustad EM, Saleski TE, Xu JH, Meinhold P, Arnold FH (2012) Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol. J Biotechnol 164:188–195. doi:10.1016/j.jbiotec.2012.08.008
Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO (2009) Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 75:6132–6141. doi:10.1128/AEM.01187-09
Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO (2009) Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 75:4315–4323. doi:10.1128/AEM.00567-09
Miller EN, Turner PC, Jarboe LR, Ingram LO (2010) Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol Lett 32:661–667. doi:10.1007/s10529-010-0209-9
Obradors N, Cabiscol E, Aguilar J, Ros J (1998) Site-directed mutagenesis studies of the metal-binding center of the iron-dependent propanediol oxidoreductase from Escherichia coli. Eur J Biochem 258:207–213
Palmqvist E, Hahn-Hagerdal B (2000) Fermentation of lignocellulosic hydrolysates. I: inhibition and detoxification. Bioresour Technol 74:17–24
Palmqvist E, Hahn-Hagerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33
Phylactides M (1997) Molecular biology series 3. Tools of molecular biology: gene cloning. Br J Hosp Med 57:49–50
Rodriguez GM, Atsumi S (2014) Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237. doi:10.1016/j.ymben.2014.07.012
Rubin EM (2008) Genomics of cellulosic biofuels. Nature 454:841–845. doi:10.1038/nature07190
Sarvari Horvath I, Franzen CJ, Taherzadeh MJ, Niklasson C, Liden G (2003) Effects of furfural on the respiratory metabolism of Saccharomyces cerevisiae in glucose-limited chemostats. Appl Environ Microbiol 69:4076–4086
Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619. doi:10.1074/jbc.M311657200
Service RF (2007) Cellulosic ethanol. Biofuel researchers prepare to reap a new harvest. Science 315:1488–1491. doi:10.1126/science.315.5818.1488
Sun Y, Cheng J (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83:1–11
van der Weijde T, Alvim Kamei CL, Torres AF, Vermerris W, Dolstra O, Visser RG, Trindade LM (2013) The potential of C4 grasses for cellulosic biofuel production. Front Plant Sci 4:107. doi:10.3389/fpls.2013.00107
Wang X, Miller EN, Yomano LP, Zhang X, Shanmugam KT, Ingram LO (2011) Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol 77:5132–5140. doi:10.1128/AEM.05008-11
Wang X, Yomano LP, Lee JY, York SW, Zheng H, Mullinnix MT, Shanmugam KT, Ingram LO (2013) Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc Natl Acad Sci USA 110:4021–4026. doi:10.1073/pnas.1217958110
Zaldivar J, Martinez A, Ingram LO (1999) Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 65:24–33
Zaldivar J, Martinez A, Ingram LO (2000) Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 68:524–530
Acknowledgments
The authors thank to Prof. Oh, Min-Kyu for kind gift of E. coli strain, DSM01. The study was partially supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015M1A5A1037196) and Advanced Production Technology Development Program, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (1201349190011). This work was also supported by the R&D Program of MOTIE/KEIT (10048350, 10049674) and the Energy Efficiency and Resources of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) grant funded by the Korea Government Ministry of Trade, Industry and Energy (20133030000300).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors claimed that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seo, HM., Jeon, JM., Lee, J.H. et al. Combinatorial application of two aldehyde oxidoreductases on isobutanol production in the presence of furfural. J Ind Microbiol Biotechnol 43, 37–44 (2016). https://doi.org/10.1007/s10295-015-1718-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1718-2