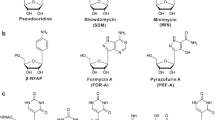
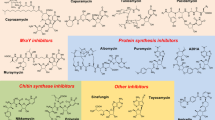
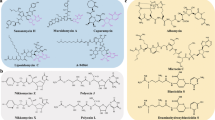
Abstract
Nucleoside antibiotics constitute an important family of microbial natural products bearing diverse bioactivities and unusual structural features. Their biosynthetic logics are unique with involvement of complex multi-enzymatic reactions leading to the intricate molecules from simple building blocks. Understanding how nature builds this family of antibiotics in post-genomic era sets the stage for rational enhancement of their production, and also paves the way for targeted persuasion of the cell factories to make artificial designer nucleoside drugs and leads via synthetic biology approaches. In this review, we discuss the recent progress and perspectives on the natural and engineered biosynthesis of nucleoside antibiotics.








Similar content being viewed by others
References
Barnard-Britson S, Chi X, Nonaka K, Spork AP, Tibrewal N, Goswami A, Pahari P, Ducho C, Rohr J, Van Lanen SG (2012) Amalgamation of nucleosides and amino acids in antibiotic biosynthesis: discovery of an l-threonine:uridine-5′-aldehyde transaldolase. J Am Chem Soc 134:18514–18517. doi:10.1021/ja308185q
Bormann C, Mohrle V, Bruntner C (1996) Cloning and heterologous expression of the entire set of structural genes for nikkomycin synthesis from Streptomyces tendae Tu901 in Streptomyces lividans. J Bacteriol 178:1216–1218
Bruntner C, Bormann C (1998) The Streptomyces tendae Tu901 l-lysine 2-aminotransferase catalyzes the initial reaction in nikkomycin D biosynthesis. Eur J Biochem 254:347–355
Chang CY, Lyu SY, Liu YC, Hsu NS, Wu CC, Tang CF, Lin KH, Ho JY, Wu CJ, Tsai MD, Li TL (2014) Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms. Angew Chem Int Ed Engl 53:1943–1948. doi:10.1002/anie.201307989
Chen H, Hubbard BK, O’Connor SE, Walsh CT (2002) Formation of beta-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem Biol 9:103–112
Chen R, Zhang H, Zhang G, Li S, Zhu Y, Liu J, Zhang C (2013) Characterizing amosamine biosynthesis in amicetin reveals AmiG as a reversible retaining glycosyltransferase. J Am Chem Soc 135:12152–12155. doi:10.1021/ja401016e
Chen W, Dai D, Wang C, Huang T, Zhai L, Deng Z (2013) Genetic dissection of the polyoxin building block-carbamoylpolyoxamic acid biosynthesis revealing the “pathway redundancy” in metabolic networks. Microb Cell Fact 12:121. doi:10.1186/1475-2859-12-121
Chen W, Huang T, He X, Meng Q, You D, Bai L, Li J, Wu M, Li R, Xie Z, Zhou H, Zhou X, Tan H, Deng Z (2009) Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J Biol Chem 284:10627–10638. doi:10.1074/jbc.M807534200
Chen W, Qu D, Zhai L, Tao M, Wang Y, Lin S, Price NP, Deng Z (2010) Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1:1093–1105. doi:10.1007/s13238-010-0127-6
Cheng L, Chen W, Zhai L, Xu D, Huang T, Lin S, Zhou X, Deng Z (2011) Identification of the gene cluster involved in muraymycin biosynthesis from Streptomyces sp. NRRL 30471. Mol BioSyst 7:920–927. doi:10.1039/c0mb00237b
Chi X, Baba S, Tibrewal N, Funabashi M, Nonaka K, Van Lanen SG (2013) The muraminomicin biosynthetic gene cluster and enzymatic formation of the 2-deoxyaminoribosyl appendage. Medchemcomm 4:239–243. doi:10.1039/C2MD20245J
Chi X, Pahari P, Nonaka K, Van Lanen SG (2011) Biosynthetic origin and mechanism of formation of the aminoribosyl moiety of peptidyl nucleoside antibiotics. J Am Chem Soc 133:14452–14459. doi:10.1021/ja206304k
Cone MC, Yin X, Grochowski LL, Parker MR, Zabriskie TM (2003) The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. ChemBioChem 4:821–828. doi:10.1002/cbic.200300583
Cunningham KG, Manson W, Spring FS, Hutchinson SA (1950) Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature 166:949
Deb Roy A, Gruschow S, Cairns N, Goss RJ (2010) Gene expression enabling synthetic diversification of natural products: chemogenetic generation of pacidamycin analogs. J Am Chem Soc 132:12243–12245. doi:10.1021/ja1060406
Delzer J, Fiedler HP, Muller H, Zahner H, Rathmann R, Ernst K, Konig WA (1984) New nikkomycins by mutasynthesis and directed fermentation. J Antibiot 37:80–82 (Tokyo)
Du D, Zhu Y, Wei J, Tian Y, Niu G, Tan H (2013) Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl Microbiol Biotechnol 97:6383–6396. doi:10.1007/s00253-013-4836-7
Feng C, Ling H, Du D, Zhang J, Niu G, Tan H (2014) Novel nikkomycin analogues generated by mutasynthesis in Streptomyces ansochromogenes. Microb Cell Fact 13:59. doi:10.1186/1475-2859-13-59
Feng J, Wu J, Dai N, Lin S, Xu HH, Deng Z, He X (2013) Discovery and characterization of BlsE, a radical S-adenosyl-l-methionine decarboxylase involved in the blasticidin S biosynthetic pathway. PLoS ONE 8:e68545. doi:10.1371/journal.pone.0068545
Feng J, Wu J, Gao J, Xia Z, Deng Z, He X (2014) Biosynthesis of the beta-methylarginine residue of peptidyl nucleoside arginomycin in Streptomyces arginensis NRRL 15941. Appl Environ Microbiol 80:5021–5027. doi:10.1128/AEM.01172-14
Fiedler HP, Kurth R, Langhärig J, Delzer J, Zähner H (1982) Nikkomycins: microbial inhibitors of chitin synthase. J Chem Technol Biotechnol 32:271–280. doi:10.1002/jctb.5030320130
Funabashi M, Baba S, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, Van Lanen SG (2010) The biosynthesis of liposidomycin-like A-90289 antibiotics featuring a new type of sulfotransferase. ChemBioChem 11:184–190. doi:10.1002/cbic.200900665
Funabashi M, Baba S, Takatsu T, Kizuka M, Ohata Y, Tanaka M, Nonaka K, Spork AP, Ducho C, Chen WC, Van Lanen SG (2013) Structure-based gene targeting discovery of sphaerimicin, a bacterial translocase I inhibitor. Angew Chem Int Ed Engl 52:11607–11611. doi:10.1002/anie.201305546
Funabashi M, Nonaka K, Yada C, Hosobuchi M, Masuda N, Shibata T, Van Lanen SG (2009) Identification of the biosynthetic gene cluster of A-500359 s in Streptomyces griseus SANK60196. J Antibiot 62:325–332. doi:10.1038/ja.2009.38 (Tokyo)
Funabashi M, Yang Z, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, Chi X, Van Lanen SG (2010) An ATP-independent strategy for amide bond formation in antibiotic biosynthesis. Nat Chem Biol 6:581–586. doi:10.1038/nchembio.393
Galgiani JN (2007) Coccidioidomycosis: changing perceptions and creating opportunities for its control. Ann N Y Acad Sci 1111:1–18. doi:10.1196/annals.1406.041
Ginj C, Ruegger H, Amrhein N, Macheroux P (2005) 3′-Enolpyruvyl-UMP, a novel and unexpected metabolite in nikkomycin biosynthesis. ChemBioChem 6:1974–1976. doi:10.1002/cbic.200500208
Gruschow S, Rackham EJ, Elkins B, Newill PL, Hill LM, Goss RJ (2009) New pacidamycin antibiotics through precursor-directed biosynthesis. ChemBioChem 10:355–360. doi:10.1002/cbic.200800575
Hector RF, Zimmer BL, Pappagianis D (1990) Evaluation of nikkomycins X and Z in murine models of coccidioidomycosis, histoplasmosis, and blastomycosis. Antimicrob Agents Chemother 34:587–593
Heitsch H, Konig WA, Decker H, Bormann C, Fiedler HP, Zahner H (1989) Metabolic products of microorganisms. 254. Structure of the new nikkomycins pseudo-Z and pseudo-J. J Antibiot (Tokyo) 42:711–717
Hiratsuka T, Suzuki H, Kariya R, Seo T, Minami A, Oikawa H (2014) Biosynthesis of the structurally unique polycyclopropanated polyketide-nucleoside hybrid jawsamycin (FR-900848). Angew Chem Int Ed Engl 53:5423–5426. doi:10.1002/anie.201402623
Isono K (1988) Nucleoside antibiotics: structure, biological activity, and biosynthesis. J Antibiot 41:1711–1739 (Tokyo)
Isono K (1991) Current progress on nucleoside antibiotics. Pharmacol Ther 52:269–286
Jackson MD, Gould SJ, Zabriskie TM (2002) Studies on the formation and incorporation of streptolidine in the biosynthesis of the peptidyl nucleoside antibiotic streptothricin F. J Org Chem 67:2934–2941
Kaysser L, Eitel K, Tanino T, Siebenberg S, Matsuda A, Ichikawa S, Gust B (2010) A new arylsulfate sulfotransferase involved in liponucleoside antibiotic biosynthesis in streptomycetes. J Biol Chem 285:12684–12694. doi:10.1074/jbc.M109.094490
Kaysser L, Lutsch L, Siebenberg S, Wemakor E, Kammerer B, Gust B (2009) Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J Biol Chem 284:14987–14996. doi:10.1074/jbc.M901258200
Kaysser L, Siebenberg S, Kammerer B, Gust B (2010) Analysis of the liposidomycin gene cluster leads to the identification of new caprazamycin derivatives. ChemBioChem 11:191–196. doi:10.1002/cbic.200900637
Kaysser L, Tang X, Wemakor E, Sedding K, Hennig S, Siebenberg S, Gust B (2011) Identification of a napsamycin biosynthesis gene cluster by genome mining. ChemBioChem 12:477–487. doi:10.1002/cbic.201000460
Kim JG, Park BK, Kim SU, Choi D, Nahm BH, Moon JS, Reader JS, Farrand SK, Hwang I (2006) Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc Natl Acad Sci USA 103:8846–8851. doi:10.1073/pnas.0602965103
Kimura K, Bugg TD (2003) Recent advances in antimicrobial nucleoside antibiotics targeting cell wall biosynthesis. Nat Prod Rep 20:252–273
Lacalle RA, Pulido D, Vara J, Zalacain M, Jimenez A (1989) Molecular analysis of the pac gene encoding a puromycin N-acetyl transferase from Streptomyces alboniger. Gene 79:375–380
Lacalle RA, Ruiz D, Jimenez A (1991) Molecular analysis of the dmpM gene encoding an O-demethyl puromycin O-methyltransferase from Streptomyces alboniger. Gene 109:55–61
Lacalle RA, Tercero JA, Jimenez A (1992) Cloning of the complete biosynthetic gene cluster for an aminonucleoside antibiotic, puromycin, and its regulated expression in heterologous hosts. EMBO J 11:785–792
Lagoja IM (2005) Pyrimidine as constituent of natural biologically active compounds. Chem Biodivers 2:1–50. doi:10.1002/cbdv.200490173
Li J, Li L, Feng C, Chen Y, Tan H (2012) Novel polyoxins generated by heterologously expressing polyoxin biosynthetic gene cluster in the sanN inactivated mutant of Streptomyces ansochromogenes. Microb Cell Fact 11:135. doi:10.1186/1475-2859-11-135
Li J, Li L, Tian Y, Niu G, Tan H (2011) Hybrid antibiotics with the nikkomycin nucleoside and polyoxin peptidyl moieties. Metab Eng 13:336–344. doi:10.1016/j.ymben.2011.01.002
Li L, Wu J, Deng Z, Zabriskie TM, He X (2013) Streptomyces lividans blasticidin S deaminase and its application in engineering a blasticidin S-producing strain for ease of genetic manipulation. Appl Environ Microbiol 79:2349–2357. doi:10.1128/AEM.03254-12
Li L, Xu Z, Xu X, Wu J, Zhang Y, He X, Zabriskie TM, Deng Z (2008) The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5′-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. ChemBioChem 9:1286–1294. doi:10.1002/cbic.200800008
Li Q, Wang L, Xie Y, Wang S, Chen R, Hong B (2013) SsaA, a member of a novel class of transcriptional regulators, controls sansanmycin production in Streptomyces sp. strain SS through a feedback mechanism. J Bacteriol 195:2232–2243. doi:10.1128/JB.00054-13
Li R, Xie Z, Tian Y, Yang H, Chen W, You D, Liu G, Deng Z, Tan H (2009) polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology 155:1819–1831. doi:10.1099/mic.0.028639-0
Liao G, Li J, Li L, Yang H, Tian Y, Tan H (2009) Selectively improving nikkomycin Z production by blocking the imidazolone biosynthetic pathway of nikkomycin X and uracil feeding in Streptomyces ansochromogenes. Microb Cell Fact 8:61. doi:10.1186/1475-2859-8-61
Liao G, Li J, Li L, Yang H, Tian Y, Tan H (2010) Cloning, reassembling and integration of the entire nikkomycin biosynthetic gene cluster into Streptomyces ansochromogenes lead to an improved nikkomycin production. Microb Cell Fact 9:6. doi:10.1186/1475-2859-9-6
Ling H, Wang G, Tian Y, Liu G, Tan H (2007) SanM catalyzes the formation of 4-pyridyl-2-oxo-4-hydroxyisovalerate in nikkomycin biosynthesis by interacting with SanN. Biochem Biophys Res Commun 361:196–201. doi:10.1016/j.bbrc.2007.07.016
Liu XY, Ruan LF, Hu ZF, Peng DH, Cao SY, Yu ZN, Liu Y, Zheng JS, Sun M (2010) Genome-wide screening reveals the genetic determinants of an antibiotic insecticide in Bacillus thuringiensis. J Biol Chem 285:39191–39200. doi:10.1074/jbc.M110.148387
Ma Z, Tao L, Bechthold A, Shentu X, Bian Y, Yu X (2014) Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl Microbiol Biotechnol 98:5051–5058. doi:10.1007/s00253-014-5573-2
Maruyama C, Toyoda J, Kato Y, Izumikawa M, Takagi M, Shin-ya K, Katano H, Utagawa T, Hamano Y (2012) A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat Chem Biol 8:791–797. doi:10.1038/nchembio.1040
McCarty RM, Bandarian V (2008) Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol 15:790–798. doi:10.1016/j.chembiol.2008.07.012
McDonald LA, Barbieri LR, Carter GT, Lenoy E, Lotvin J, Petersen PJ, Siegel MM, Singh G, Williamson RT (2002) Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J Am Chem Soc 124:10260–10261
Mohrle V, Roos U, Bormann C (1995) Identification of cellular proteins involved in nikkomycin production in Streptomyces tendae Tu901. Mol Microbiol 15:561–571
Moon M, Van Lanen SG (2010) Characterization of a dual specificity aryl acid adenylation enzyme with dual function in nikkomycin biosynthesis. Biopolymers 93:791–801. doi:10.1002/bip.21479
Muramatsu Y, Arai M, Sakaida Y, Takamatsu Y, Miyakoshi S, Inukai M (2006) Studies on novel bacterial translocase I inhibitors, A-500359 s. V. Enhanced production of capuramycin and A-500359 A in Streptomyces griseus SANK 60196. J Antibiot 59:601–606. doi:10.1038/ja.2006.81 (Tokyo)
Muramatsu Y, Ohnuki T, Ishii MM, Kizuka M, Enokita R, Miyakoshi S, Takatsu T, Inukai M (2004) A-503083 A, B, E and F, novel inhibitors of bacterial translocase I, produced by Streptomyces sp. SANK 62799. J Antibiot 57:639–646 (Tokyo)
Niu G, Li L, Wei J, Tan H (2013) Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chem Biol 20:34–44. doi:10.1016/j.chembiol.2012.10.017
Niu G, Liu G, Tian Y, Tan H (2006) SanJ, an ATP-dependent picolinate-CoA ligase, catalyzes the conversion of picolinate to picolinate-CoA during nikkomycin biosynthesis in Streptomyces ansochromogenes. Metab Eng 8:183–195. doi:10.1016/j.ymben.2005.12.002
Niu G, Tan H (2014) Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends Microbiol. doi:10.1016/j.tim.2014.10.007
Osada H, Isono K (1985) Mechanism of action and selective toxicity of ascamycin, a nucleoside antibiotic. Antimicrob Agents Chemother 27:230–233
Pan Y, Lu C, Dong H, Yu L, Liu G, Tan H (2013) Disruption of rimP-SC, encoding a ribosome assembly cofactor, markedly enhances the production of several antibiotics in Streptomyces coelicolor. Microb Cell Fact 12:65. doi:10.1186/1475-2859-12-65
Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ (2010) Pacidamycin biosynthesis: identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. ChemBioChem 11:1700–1709. doi:10.1002/cbic.201000200
Ragab AE, Gruschow S, Tromans DR, Goss RJ (2011) Biogenesis of the unique 4′,5′-dehydronucleoside of the uridyl peptide antibiotic pacidamycin. J Am Chem Soc 133:15288–15291. doi:10.1021/ja206163j
Rosemeyer H (2004) The chemodiversity of purine as a constituent of natural products. Chem Biodivers 1:361–401. doi:10.1002/cbdv.200490033
Saugar I, Sanz E, Rubio MA, Espinosa JC, Jimenez A (2002) Identification of a set of genes involved in the biosynthesis of the aminonucleoside moiety of antibiotic A201A from Streptomyces capreolus. Eur J Biochem 269:5527–5535
Stenland CJ, Lis LG, Schendel FJ, Hahn NJ, Smart MA, Miller AL, von Keitz MG, Gurvich VJ (2013) A practical and scalable manufacturing process for an anti-fungal agent, Nikkomycin Z. Org Process Res Dev 17:265–272. doi:10.1021/op3003294
Tang X, Eitel K, Kaysser L, Kulik A, Grond S, Gust B (2013) A two-step sulfation in antibiotic biosynthesis requires a type III polyketide synthase. Nat Chem Biol 9:610–615. doi:10.1038/nchembio.1310
Tercero JA, Espinosa JC, Jimenez A (1998) Expression of the Streptomyces alboniger pur cluster in Streptomyces lividans is dependent on the bldA-encoded tRNALeu. FEBS Lett 421:221–223
Tercero JA, Espinosa JC, Lacalle RA, Jimenez A (1996) The biosynthetic pathway of the aminonucleoside antibiotic puromycin, as deduced from the molecular analysis of the pur cluster of Streptomyces alboniger. J Biol Chem 271:1579–1590
Tokiwano T, Watanabe H, Seo T, Oikawa H (2008) Unprecedented biological cyclopropanation in the biosynthesis of FR-900848. Chem Commun (Camb):6016-6018. doi:10.1039/b809610d
Venci D, Zhao G, Jorns MS (2002) Molecular characterization of NikD, a new flavoenzyme important in the biosynthesis of nikkomycin antibiotics. Biochemistry 41:15795–15802
Walsh CT, Zhang W (2011) Chemical logic and enzymatic machinery for biological assembly of peptidyl nucleoside antibiotics. ACS Chem Biol 6:1000–1007. doi:10.1021/cb200284p
Winn M, Goss RJ, Kimura K, Bugg TD (2010) Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep 27:279–304. doi:10.1039/b816215h
Wyszynski FJ, Hesketh AR, Bibb MJ, Davis BG (2010) Dissecting tunicamycin biosynthesis by genome mining: cloning and heterologous expression of a minimal gene cluster. Chem Sci 1:9. doi:10.1039/C0SC00325E
Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, Bibb MJ, Lee SJ, Davies GJ, Davis BG (2012) Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nat Chem 4:539–546. doi:10.1038/nchem.1351
Xie Y, Cai Q, Ren H, Wang L, Xu H, Hong B, Wu L, Chen R (2014) NRPS substrate promiscuity leads to more potent antitubercular sansanmycin analogues. J Nat Prod 77:1744–1748. doi:10.1021/np5001494
Xie Z, Niu G, Li R, Liu G, Tan H (2007) Identification and characterization of sanH and sanI involved in the hydroxylation of pyridyl residue during nikkomycin biosynthesis in Streptomyces ansochromogenes. Curr Microbiol 55:537–542. doi:10.1007/s00284-007-9028-1
Xu D, Liu G, Cheng L, Lu X, Chen W, Deng Z (2013) Identification of Mur34 as the novel negative regulator responsible for the biosynthesis of muraymycin in Streptomyces sp. NRRL30471. PLoS ONE 8:e76068. doi:10.1371/journal.pone.0076068
Yang Z, Chi X, Funabashi M, Baba S, Nonaka K, Pahari P, Unrine J, Jacobsen JM, Elliott GI, Rohr J, Van Lanen SG (2011) Characterization of LipL as a non-heme, Fe(II)-dependent alpha-ketoglutarate:uMP dioxygenase that generates uridine-5′-aldehyde during A-90289 biosynthesis. J Biol Chem 286:7885–7892. doi:10.1074/jbc.M110.203562
Yang Z, Funabashi M, Nonaka K, Hosobuchi M, Shibata T, Pahari P, Van Lanen SG (2010) Functional and kinetic analysis of the phosphotransferase CapP conferring selective self-resistance to capuramycin antibiotics. J Biol Chem 285:12899–12905. doi:10.1074/jbc.M110.104141
Yoshida M, Ezaki M, Hashimoto M, Yamashita M, Shigematsu N, Okuhara M, Kohsaka M, Horikoshi K (1990) A novel antifungal antibiotic, FR-900848. I. Production, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 43:748–754
Zeng Y, Kulkarni A, Yang Z, Patil PB, Zhou W, Chi X, Van Lanen S, Chen S (2012) Biosynthesis of albomycin delta(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem Biol 7:1565–1575. doi:10.1021/cb300173x
Zeng Y, Roy H, Patil PB, Ibba M, Chen S (2009) Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974. Antimicrob Agents Chemother 53:4619–4627. doi:10.1128/AAC.00782-09
Zhai L, Lin S, Qu D, Hong X, Bai L, Chen W, Deng Z (2012) Engineering of an industrial polyoxin producer for the rational production of hybrid peptidyl nucleoside antibiotics. Metab Eng 14:388–393. doi:10.1016/j.ymben.2012.03.006
Zhang G, Zhang H, Li S, Xiao J, Zhu Y, Niu S, Ju J, Zhang C (2012) Characterization of the amicetin biosynthesis gene cluster from Streptomyces vinaceusdrappus NRRL 2363 implicates two alternative strategies for amide bond formation. Appl Environ Microbiol 78:2393–2401. doi:10.1128/AEM.07185-11
Zhang W, Ntai I, Bolla ML, Malcolmson SJ, Kahne D, Kelleher NL, Walsh CT (2011) Nine enzymes are required for assembly of the pacidamycin group of peptidyl nucleoside antibiotics. J Am Chem Soc 133:5240–5243. doi:10.1021/ja2011109
Zhang W, Ntai I, Kelleher NL, Walsh CT (2011) tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc Natl Acad Sci USA 108:12249–12253. doi:10.1073/pnas.1109539108
Zhang W, Ostash B, Walsh CT (2010) Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc Natl Acad Sci USA 107:16828–16833. doi:10.1073/pnas.1011557107
Zhao C, Qi J, Tao W, He L, Xu W, Chan J, Deng Z (2014) Characterization of biosynthetic genes of ascamycin/dealanylascamycin featuring a 5′-O-sulfonamide moiety in Streptomyces sp. JCM9888. PLoS ONE 9:e114722. doi:10.1371/journal.pone.0114722
Zhao G, Wu G, Zhang Y, Liu G, Han T, Deng Z, He X (2014) Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry. Nucleic Acids Res 42:8115–8124. doi:10.1093/nar/gku486
Zhu Q, Li J, Ma J, Luo M, Wang B, Huang H, Tian X, Li W, Zhang S, Zhang C, Ju J (2012) Discovery and engineered overproduction of antimicrobial nucleoside antibiotic A201A from the deep-sea marine actinomycete Marinactinospora thermotolerans SCSIO 00652. Antimicrob Agents Chemother 56:110–114. doi:10.1128/AAC.05278-11
Acknowledgments
We thank Dr. Neil Price (NCAUR-USDA, Peoria, Il, USA) for editorial comments on the manuscript. This work was supported by Grants 973 (2012CB721004) from the Ministry of Science and Technology, the National Science Foundation of China (31270100, 31070027), Wuhan Youth Chenguang Program of Science and Technology (201507040401018), the Open Funding Project of the State Key Laboratory of Bioreactor Engineering, and the Open Funding Project of the State Key Laboratory of Microbial Metabolism.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Natural Product Discovery and Development in the Genomic Era. Dedicated to Professor Satoshi Ōmura for his numerous contributions to the field of natural products.
Rights and permissions
About this article
Cite this article
Chen, W., Qi, J., Wu, P. et al. Natural and engineered biosynthesis of nucleoside antibiotics in Actinomycetes . J Ind Microbiol Biotechnol 43, 401–417 (2016). https://doi.org/10.1007/s10295-015-1636-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1636-3