Abstract
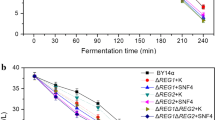
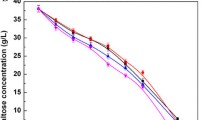
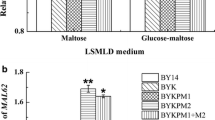
Dough-leavening ability is one of the main aspects considered when selecting a baker’s yeast strain for baking industry. Generally, modification of maltose metabolic pathway and known regulatory networks of maltose metabolism were used to increase maltose metabolism to improve leavening ability in lean dough. In this study, we focus on the effects of PGM2 (encoding for the phosphoglucomutase) and SNR84 (encoding for the H/ACA snoRNA) that are not directly related to both the maltose metabolic pathway and known regulatory networks of maltose metabolism on the leavening ability of baker’s yeast in lean dough. The results show that the modifications on PGM2 and/or SNR84 are effective ways in improving leavening ability of baker’s yeast in lean dough. Deletion of PGM2 decreased cellular glucose-1-phosphate and overexpression of SNR84 increased the maltose permease activity. These changes resulted in 11, 19 and 21 % increases of the leavening ability for PGM2 deletion, SNR84 overexpression and SNR84 overexpression combining deleted PGM2, respectively.




Similar content being viewed by others
References
Alves-Araújo C, Pacheco A, Almeida MJ, Spencer-Martins I, Leão C, Sousa MJ (2007) Sugar utilization patterns and respiro-fermentative metabolism in the baker’s yeast Torulaspora delbrueckii. Microbiology 153:898–904
Angelov AI, Karadjov GI, Roshkova ZG (1996) Strains selection of baker’s yeast with improved technological properties. Food Res Int 29:235–239
Bell PJ, Higgins VJ, Attfield PV (2001) Comparison of fermentative capacities of industrial baking and wild-type yeasts of the species Saccharomyces cerevisiae in different sugar media. Lett Appl Microbiol 32:224–229
Bro C, Knudsen S, Regenberg B, Olsson L, Nielsen J (2005) Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol 71:6465–6472
Brown JW, Clark GP, Leader DJ, Simpson CG, Lowe T (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA 7:1817–1832
Campelo AF, Belo I (2004) Fermentative capacity of baker’s yeast exposed to hyperbaric stress. Biotechnol Lett 26:1237–1240
Chen Y, Liu Q, Chen X, Wu J, Xie J, Guo T, Zhu C, Ying H (2014) Control of glycolytic flux in directed biosynthesis of uridine-phosphoryl compounds through the manipulation of ATP availability. Appl Microbiol Biotechnol 98:6621–6632
Garcia Sanchez R, Hahn-Hägerdal B, Gorwa-Grauslund MF (2010) PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae. Microb Cell Fact 9:40
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30:e23
Hauf J, Zimmermann FK, Müller S (2000) Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enzyme Microb Technol 26:688–698
Higgins VJ, Braidwood M, Bell P, Bissinger P, Dawes IW, Attfield PV (1999) Genetic evidence that high noninduced maltase and maltose permease activities, governed by MALx3-encoded transcriptional regulators, determine efficiency of gas production by baker’s yeast in unsugared dough. Appl Environ Microbiol 65:680–685
Hirasawa R, Yokoigawa K (2001) Leavening ability of baker’s yeast exposed to hyperosmotic media. FEMS Microbiol Lett 194:159–162
Houghton-Larsen J, Brandt A (2006) Fermentation of high concentrations of maltose by Saccharomyces cerevisiae is limited by the COMPASS methylation complex. Appl Environ Microbiol 72:7176–7182
Howard SC, Deminoff SJ, Herman PK (2006) Increased phosphoglucomutase activity suppresses the galactose growth defect associated with elevated levels of Ras signaling in S. cerevisiae. Curr Genet 49:1–6
Hu Z, Nehlin JO, Ronne H, Michels CA (1995) MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr Genet 28:258–266
Jiang TX, Xiao DG, Gao Q (2008) Characterisation of maltose metabolism in lean dough by lagging and non-lagging baker’s yeast strains. Ann Microbiol 58:655–660
Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145–148
Klein CJL, Olsson L, Rønnow B, Mikkelsen JD, Nielsen J (1996) Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl Environ Microbiol 62:4441–4449
Klein CJL, Olsson L, Ronnow B, Mikkelsen JD, Nielsen J (1997) Glucose and Maltose Metabolism in MIG1-disrupted and MAL-constitutive Strains of Saccharomyces cerevisiae. Food Technol Biotechnol 35:287–292
Lee KS, Hong ME, Jung SC, Ha SJ, Yu BJ, Koo HM, Park SM, Seo JH, Kweon DH, Park JC, Jin YS (2011) Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng 108:621–631
Lian J, Si T, Nair NU, Zhao H (2014) Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng 24:139–149
Lilly M, Lambrechts MG, Pretorius IS (2000) Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl Environ Microbiol 66:744–753
Lin X, Zhang CY, Bai XW, Song HY, Xiao DG (2014) Effects of MIG1, TUP1 and SSN6 deletion on maltose metabolism and leavening ability of baker’s yeast in lean dough. Microb Cell Fact 13:93
Lu J, Dong J, Wu D, Chen Y, Guo X, Shi Y, Sun X, Xiao D (2012) Construction of recombinant industrial brewer’s yeast with lower diacetyl production and proteinase A activity. Eur Food Res Technol 235:951–961
Ni J, Tien AL, Fournier MJ (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565–573
Oda Y, Ouchi K (1990) Effect of chromosome loss on the leavening ability of Saccharomyces cerevisiae in dough. Curr Genet 18:401–403
Oh D, Hopper JE (1990) Transcription of a yeast phosphoglucomutase isozyme gene is galactose inducible and glucose repressible. Mol Cell Biol 10:1415–1422
Randez-Gil F, Córcoles-Sáez I, Prieto JA (2013) Genetic and phenotypic characteristics of baker’s yeast: relevance to baking. Annu Rev Food Sci Technol 4:191–214
Randez-Gil F, Sanz P, Prieto JA (1999) Engineering baker’s yeast: room for improvement. Trends Biotechnol 17:237–244
Salema-Oom M, De Sousa HR, Assunçao M, Gonçalves P, Spencer-Martins I (2011) Derepression of a baker’s yeast strain for maltose utilization is associated with severe deregulation of HXT gene expression. J Appl Microbiol 110:364–374
Sasano Y, Haitani Y, Hashida K, Oshiro S, Shima J, Takagi H (2013) Improvement of fermentation ability under baking-associated stress conditions by altering the POG1 gene expression in baker’s yeast. Int J Food Microbiol 165:241–245
Schattner P, Decatur WA, Davis CA, Ares M Jr, Fournier MJ, Lowe TM (2004) Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res 32:4281–4296
Schüller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160
Sun X, Zhang C, Dong J, Wu M, Zhang Y, Xiao D (2012) Enhanced leavening properties of baker’s yeast overexpressing MAL62 with deletion of MIG1 in lean dough. J Ind Microbiol Biotechnol 39:1533–1539
Tycowski KT, Steitz JA (2001) Non-coding snoRNA host genes in Drosophila: expression strategies for modification guide snoRNAs. Eur J Cell Biol 80:119–125
Acknowledgments
The current study was financially supported by the National Natural Science Foundation of China (31171730), the National High Technology Research and Development Program of China (2013AA102106), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT1166).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lin, X., Zhang, CY., Bai, XW. et al. Enhanced leavening ability of baker’s yeast by overexpression of SNR84 with PGM2 deletion. J Ind Microbiol Biotechnol 42, 939–948 (2015). https://doi.org/10.1007/s10295-015-1618-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1618-5