Abstract
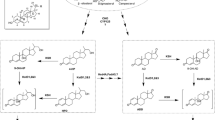
Mycobacterium neoaurum TCCC 11028 (MNR) and M. neoaurum TCCC 11028 M3 (MNR M3) significantly differ in the ratio of androst-1,4-diene-3,17-dione (ADD) to androst-4-ene-3,17-dione (AD) produced. The large fluctuations are related to the dehydrogenation activity of 3-ketosteroid-Δ1-dehydrogenase (KsdD). Analysis of the primary structure of KsdD showed that the Ser-138 of KsdD-MNR changed to Leu-138 of KsdD-MNR M3 because of C413T in the ksdD gene. This phenomenon directly affected KsdD activity. The effect of the primary structure of KsdD on dehydrogenation activity was confirmed through exogenous expression. Whole-cell transformation initially revealed that KsdD-MNR showed a higher dehydrogenation activity than KsdD-MNR M3. Then, ksdD gene replacement strain was constructed by homologous recombination. The results of steroid transformation experiments showed that the ability of the MNR M3ΔksdD::ksdD-MNR strain to produce ADD was improved and it returned to the similar level of the MNR strain. This result indicated that the ADD/AD ratio of the two M. neoaurum strains was influenced by the difference in ksdD. The mechanism by which residue mutations alter enzyme activity may be connected with the crystal structure of KsdD from Rhodococcus erythropolis SQ1. As a key amino acid residue in the active center position, Ser-138 played an important role in maintaining the active center in the hydrophobic environment of KsdD. This study may serve as a basis for future studies on the structural analysis and catalytic mechanism of dehydrogenase.




Similar content being viewed by others
References
Adham NZ, EI-Hady AA, Naim N (2003) Biochemical studies on the microbial Δ1-dehydrogenation of cortisol by Pseudomonas fluorescens. Process Biochem 38(6):897–902
Bragin EY, Shtratnikova VY, Dovbnya DV, Schelkunov MI, Pekov YA, Malakho SG, Egorova OV, Ivashina TV, Sokolov SL, Ashapkin VV, Donova MV (2013) Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. Strains. J Steroid Biochem 138:41–53
Brzostek A, Sliwinski T, Rumijowska-Galewicz A, Korycka-Machala M, Dziadek J (2005) Identification and targeted disruption of the gene encoding the main 3-ketosteroid dehydrogenase in Mycobacterium smegmatis. Microbiology 151(7):2393–2402
Donova MV (2007) Transformation of steroids by actinobacteria: a review. Appl Biochem Micro 43(1):1–14
Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS (2003) Microbial conversion of steroid compounds: recent developments. Enzyme Microb Tech 32(6):688–705
Fokina VV, Karpov AV, Sidorov IA, Andrjushina VA, Arinbasarova AY (1997) The influence of β-cyclodextrin on the kinetics of 1-en-dehydrogenation of 6α-methylhydro cortisone by Arthrobacter globiformis cells. Appl Microbiol Biot 47:645–649
Fujii C, Morii S, Kadode M, Sawamoto S, Iwami M, Itagaki E (1999) Essential tyrosine residues in 3-ketosteroid-Δ1-dehydrogenase from Rhodococcus rhodochrous. J Biochem 126(4):662–667
Geize R, Hessels GI, Grommen R, Vrijbloed JW, Meijden P, Dijkhuizen L (2000) Targeted disruption of the kstD gene encoding a 3-ketosteroid Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl Environ Microb 66(5):2029–2036
Geize R, Hessels GI, Dijkhuizen L (2002) Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid Δ1-dehydrogenase isoenzyme. Microbiology 148(10):3285–3292
Knol J, Bodewits K, Hessels GI, Dijkhuizen L, Geize R (2008) 3-Keto-5α-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem J 410(2):339–346
Li Y, Lu F, Sun T, Du L (2007) Expression of ksdD gene encoding 3-ketosteroid-Δ1-dehydrogenase from Arthrobacter simplex in Bacillus subtilis. Lett Appl Microbiol 44(5):563–568
Matsushita H, Itagaki E (1992) Essential histidine residue in 3-ketosteroid-Δ1-dehydrogenase. J Biochem 111:594–599
Molnar I, Choi KP, Yamashita M, Murooka Y (1995) Molecular cloning, expression in Streptomyces livdans, and analysis of a gene cluster from Arthrobacter simplex encoding 3-ketosteroid-Δ1-dehydrogenase, 3-ketosteroid-Δ5-isomerase and a hypothetical regulatory protein. Mol Microbiol 15(5):895–905
Oosterwijk N, Knol J, Dijkhuizen L, Geize R, Dijkstra BW (2012) Structure and catalytic mechanism of 3-ketosteroid-Δ4-(5α)-dehydrogenase from Rhodococcus jostii RHA1 genome. J Biol Chem 287(37):30975–30983
Oosterwijk N, Knol J, Dijkhuizen L, Geize R, Dijkstra BW (2011) Cloning, overexpression, purification, crystallization and preliminary X-ray analysis of 3-ketosteroid Δ4-(5α)-dehydrogenase from Rhodococcus jostii RHA1. Acta Crystallogr F 67:1269–1273
Parish T, Stoker NG (2000) Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(8):1969–1975
Plesiat P, Grandguillot M, Harayama S, Vragar S, Michel-Briand Y (1991) Cloning, sequencing, and expression of the Pseudomonas testosteroni gene encoding 3-oxosteroid Δ1-dehydrogenase. J Bacteriol 173(22):7219–7227
Rohman A, Oosterwijk N, Thunnissen AMW, Dijkstra BW (2013) Crystal structure and site-directed mutagenesis of 3-ketosteroid-Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. J Biol Chem 288(49):35559–35568
Shen YB, Wang M, Li HN, Wang YB, Luo JM (2012) Influence of hydroxypropyl-β-cyclodextrin on phytosterol biotransformation by different strains of Mycobacterium neoaurum. J Ind Microbiol Biot 39(9):1253–1259
Uhía I, Galán B, Kendall SL, Stoker NG, García JL (2012) Cholesterol metabolism in Mycobacterium smegmatis. Env Microbiol Rep 4(2):168–182
Wei W, Wang FQ, Fan SY, Wei DZ (2010) Inactivation and augmentation of the primary 3-ketosteroid-Δ1-dehydrogenase in Mycobacterium neoaurum NwIB-01: biotransformation of soybean phytosterols to 4-androstene- 3,17-dione or 1,4-androstadiene-3,17-dione. Appl Environ Microb 76(13):4578–4582
Yao K, Xu LQ, Wang FQ, Wei DZ (2014) Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab Eng 24:181–191
Zhang CG, Wang FQ, Wei DZ (2011) The clone and expression of 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum NwIB-01. Chem Bioeng 28(7):39–43 (In Chinese)
Acknowledgments
This work was supported by the National High Technology Research and Development of China (2011AA02A211), the National Natural Science Foundation of China (21276196, 21406167), and Key Project of Chinese Ministry of Education (213004A).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xie, R., Shen, Y., Qin, N. et al. Genetic differences in ksdD influence on the ADD/AD ratio of Mycobacterium neoaurum . J Ind Microbiol Biotechnol 42, 507–513 (2015). https://doi.org/10.1007/s10295-014-1577-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1577-2